Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A
- PMID: 15096578
- PMCID: PMC407823
- DOI: 10.1093/nar/gkh540
Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A
Abstract
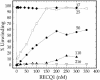
RecQ helicases are required for the maintenance of genome stability. Characterization of the substrate specificity and identification of the binding partners of the five human RecQ helicases are essential for understanding their function. In the present study, we have developed an efficient baculovirus expression system that allows us to obtain milligram quantities of recombinant RECQ1. Our gel filtration and dynamic light scattering experiments show that RECQ1 has an apparent molecular mass of 158 kDa and a hydrodynamic radius of 5.4 +/- 0.6 nm, suggesting that RECQ1 forms dimers in solution. The oligomeric state of RECQ1 remains unchanged upon binding to a single-stranded (ss)DNA fragment of 50 nt. We show that RECQ1 alone is able to unwind short DNA duplexes (<110 bp), whereas considerably longer substrates (501 bp) can be unwound only in the presence of human replication protein A (hRPA). The same experiments with Escherichia coli SSB show that RECQ1 is specifically stimulated by hRPA. However, hRPA does not affect the ssDNA-dependent ATPase activity of RECQ1. In addition, our far western, ELISA and co-immunoprecipitation experiments demonstrate that RECQ1 physically interacts with the 70 kDa subunit of hRPA and that this interaction is not mediated by DNA.
Figures



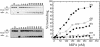



Similar articles
-
Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A.J Biol Chem. 2003 Jan 17;278(3):1424-32. doi: 10.1074/jbc.M209407200. Epub 2002 Nov 4. J Biol Chem. 2003. PMID: 12419808
-
Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity.J Biol Chem. 2000 Aug 4;275(31):23500-8. doi: 10.1074/jbc.M001557200. J Biol Chem. 2000. PMID: 10825162
-
Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1.J Biol Chem. 2005 Jul 29;280(30):28072-84. doi: 10.1074/jbc.M500264200. Epub 2005 May 16. J Biol Chem. 2005. PMID: 15899892
-
Distinct roles of RECQ1 in the maintenance of genomic stability.DNA Repair (Amst). 2010 Mar 2;9(3):315-24. doi: 10.1016/j.dnarep.2009.12.010. Epub 2010 Jan 12. DNA Repair (Amst). 2010. PMID: 20061189 Free PMC article. Review.
-
Non-hexameric DNA helicases and translocases: mechanisms and regulation.Nat Rev Mol Cell Biol. 2008 May;9(5):391-401. doi: 10.1038/nrm2394. Nat Rev Mol Cell Biol. 2008. PMID: 18414490 Review.
Cited by
-
Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition.Nat Struct Mol Biol. 2013 Mar;20(3):347-54. doi: 10.1038/nsmb.2501. Epub 2013 Feb 10. Nat Struct Mol Biol. 2013. PMID: 23396353 Free PMC article.
-
Replication protein A stimulates the Werner syndrome protein branch migration activity.J Biol Chem. 2009 Dec 11;284(50):34682-91. doi: 10.1074/jbc.M109.049031. Epub 2009 Oct 7. J Biol Chem. 2009. PMID: 19812417 Free PMC article.
-
RECQ1 possesses DNA branch migration activity.J Biol Chem. 2008 Jul 18;283(29):20231-42. doi: 10.1074/jbc.M801582200. Epub 2008 May 21. J Biol Chem. 2008. PMID: 18495662 Free PMC article.
-
The p63 protein isoform ΔNp63α modulates Y-box binding protein 1 in its subcellular distribution and regulation of cell survival and motility genes.J Biol Chem. 2012 Aug 31;287(36):30170-80. doi: 10.1074/jbc.M112.349951. Epub 2012 Jul 11. J Biol Chem. 2012. PMID: 22787154 Free PMC article.
-
Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain.Nucleic Acids Res. 2008 Nov;36(19):6295-308. doi: 10.1093/nar/gkn609. Epub 2008 Oct 3. Nucleic Acids Res. 2008. PMID: 18835853 Free PMC article.
References
-
- Wu L. and Hickson,I.D. (2001) Molecular biology. DNA ends ReQ-uire attention. Science, 292, 229–230. - PubMed
-
- Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. - PubMed
-
- Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. - PubMed
-
- Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. - PubMed
-
- Kitao S., Lindor,N.M., Shiratori,M., Furuichi,Y. and Shimamoto,A. (1999) Rothmund-Thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics, 61, 268–276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

