Stochastic model of protein-protein interaction: why signaling proteins need to be colocalized
- PMID: 15096590
- PMCID: PMC404064
- DOI: 10.1073/pnas.0401314101
Stochastic model of protein-protein interaction: why signaling proteins need to be colocalized
Abstract
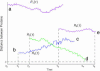
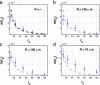
Colocalization of proteins that are part of the same signal transduction pathway via compartmentalization, scaffold, or anchor proteins is an essential aspect of the signal transduction system in eukaryotic cells. If interaction must occur via free diffusion, then the spatial separation between the sources of the two interacting proteins and their degradation rates become primary determinants of the time required for interaction. To understand the role of such colocalization, we create a mathematical model of the diffusion based protein-protein interaction process. We assume that mRNAs, which serve as the sources of these proteins, are located at different positions in the cytoplasm. For large cells such as Drosophila oocytes we show that if the source mRNAs were at random locations in the cell rather than colocalized, the average rate of interactions would be extremely small, which suggests that localization is needed to facilitate protein interactions and not just to prevent cross-talk between different signaling modules.
Figures


References
-
- Scott, J. D. & Pawson, T. (2000) Sci. Am. 282 (6), 72-79. - PubMed
-
- Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717-728. - PubMed
-
- Velculescu, V. E., Madden, S. L., Zhang, L., Lash, A. E., Yu, J., Rago, C., Lal, A., Wang, C. J., Beaudry, G. A., Ciriello, K. M., et al. (1999) Nat. Genet. 23, 387-388. - PubMed
-
- Blake, W. J., Ern, M. K. A., Cantor, C. R. & Collins, J. J. (2003) Nature 422, 633-637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

