An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp
- PMID: 15096594
- PMCID: PMC419640
- DOI: 10.1073/pnas.0401634101
An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp
Abstract
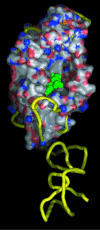
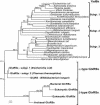
The product of the Escherichia coli yadB gene is homologous to the N-terminal part of bacterial glutamyl-tRNA synthetases (GluRSs), including the Rossmann fold with the acceptor-binding domain and the stem-contact fold. This GluRS-like protein, which lacks the anticodon-binding domain, does not use tRNA(Glu) as substrate in vitro nor in vivo, but aminoacylates tRNA(Asp) with glutamate. The yadB gene is expressed in wild-type E. coli as an operon with the dksA gene, which encodes a protein involved in the general stress response by means of its action at the translational level. The fate of the glutamylated tRNA(Asp) is not known, but its incapacity to bind elongation factor Tu suggests that it is not involved in ribosomal protein synthesis. Genes homologous to yadB are present only in bacteria, mostly in Proteobacteria. Sequence alignments and phylogenetic analyses show that the YadB proteins form a distinct monophyletic group related to the bacterial and organellar GluRSs (alpha-type GlxRSs superfamily) with ubiquitous function as suggested by the similar functional properties of the YadB homologue from Neisseria meningitidis.
Figures



Comment in
-
Turning tRNA upside down: When aminoacylation is not a prerequisite to protein synthesis.Proc Natl Acad Sci U S A. 2004 May 18;101(20):7493-4. doi: 10.1073/pnas.0402276101. Epub 2004 May 11. Proc Natl Acad Sci U S A. 2004. PMID: 15138304 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

