Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease
- PMID: 15096615
- PMCID: PMC404055
- DOI: 10.1073/pnas.0401031101
Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease
Abstract
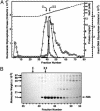
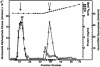
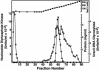
Escherichia coli nucleoside diphosphate kinase (Ndk) catalyzes ATP-dependent synthesis of ribo- and deoxyribonucleoside triphosphates from the cognate diphosphate precursor. Recently, the Ndk polypeptide was reported to be a multifunctional base excision repair nuclease that processed uracil residues in DNA by acting sequentially as a uracil-DNA glycosylase inhibitor protein (Ugi)-sensitive uracil-DNA glycosylase, an apurinic/apyrimidiniclyase, and a 3'-phosphodiesterase [Postel, E. H. & Abramczyk, B. M. (2003) Proc. Natl. Acad. Sci. USA 100, 13247-13252]. Here we demonstrate that the E. coli Ndk polypeptide lacked detectable uracil-DNA glycosylase activity and, hence, was incapable of acting as a uracil-processing DNA repair nuclease. This finding was based on the following observations: (i) uracil-DNA glycosylase activity did not copurify with Ndk activity; (ii) Ndk purified from E. coli ung(-) cells showed no detectable uracil-DNA glycosylase activity; and (iii) Ndk failed to bind to a Ugi-Sepharose affinity column that tightly bound E. coli uracil-DNA glycosylase (Ung). Collectively, these observations demonstrate that the E. coli Ndk polypeptide does not possess inherent uracil-DNA glycosylase activity.
Figures
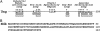


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

