Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae
- PMID: 15096616
- PMCID: PMC404124
- DOI: 10.1073/pnas.0401090101
Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae
Abstract
Viroids, subviral noncoding RNAs, replicate, move, and incite diseases in plants. Viroids replicate through a rolling-circle mechanism in which oligomeric RNAs of one or both polarities are cleaved and ligated into the circular monomers. Attempts to transmit viroids to Arabidopsis have failed for unknown reasons. To tackle this question, Arabidopsis was transformed with cDNAs expressing dimeric (+) transcripts of representative species of the families Pospiviroidae and Avsunviroidae, which replicate in the nucleus and the chloroplast, respectively. Correct processing to the circular (+) monomers was always observed, demonstrating that Arabidopsis has the appropriate RNase and RNA ligase. Northern blot hybridization also revealed the multimeric (-) RNAs of Citrus exocortis viroid and Hop stunt viroid (HSVd) of the family Pospiviroidae, but not of Avocado sunblotch viroid of the family Avsunviroidae, showing that the first RNA-RNA transcription of the rolling-circle mechanism occurs in Arabidopsis for the two nuclear viroids and that their multimeric (-) RNAs remain unprocessed as in typical hosts. Moreover, transgenic Arabidopsis expressing HSVd dimeric (-) transcripts accumulated the circular (+) monomers, although at low levels, together with the unprocessed primary transcript that served as the template for the second RNA-RNA transcription. Agroinoculation of Arabidopsis with the dimeric (+) Citrus exocortis viroid, HSVd, and Coleus blumei viroid 1 cDNAs showed that these viroids could not move to distal plant parts, in contrast with the situation observed in their experimental hosts. Therefore, deficiencies in movement or low replication appear to be the factors limiting infectivity of some viroids in Arabidopsis.
Figures

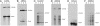
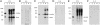

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

