Informational complexity and functional activity of RNA structures
- PMID: 15099096
- PMCID: PMC5042360
- DOI: 10.1021/ja031504a
Informational complexity and functional activity of RNA structures
Abstract
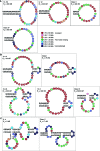
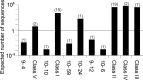
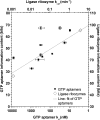
Very little is known about the distribution of functional DNA, RNA, and protein molecules in sequence space. The question of how the number and complexity of distinct solutions to a particular biochemical problem varies with activity is an important aspect of this general problem. Here we present a comparison of the structures and activities of eleven distinct GTP-binding RNAs (aptamers). By experimentally measuring the amount of information required to specify each optimal binding structure, we show that defining a structure capable of 10-fold tighter binding requires approximately 10 additional bits of information. This increase in information content is equivalent to specifying the identity of five additional nucleotide positions and corresponds to an approximately 1000-fold decrease in abundance in a sample of random sequences. We observe a similar relationship between structural complexity and activity in a comparison of two catalytic RNAs (ribozyme ligases), raising the possibility of a general relationship between the complexity of RNA structures and their functional activity. Describing how information varies with activity in other heteropolymers, both biological and synthetic, may lead to an objective means of comparing their functional properties. This approach could be useful in predicting the functional utility of novel heteropolymers.
Figures


References
-
- Lehman N.; Donne M. D.; West M.; Dewey T. G The genotypic landscape during in vitro evolution of a catalytic RNA: implications for phenotypic buffering. J. Mol. Evol. 2000, 50, 481–490. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

