BioBuilder as a database development and functional annotation platform for proteins
- PMID: 15099404
- PMCID: PMC406495
- DOI: 10.1186/1471-2105-5-43
BioBuilder as a database development and functional annotation platform for proteins
Abstract
Background: The explosion in biological information creates the need for databases that are easy to develop, easy to maintain and can be easily manipulated by annotators who are most likely to be biologists. However, deployment of scalable and extensible databases is not an easy task and generally requires substantial expertise in database development.
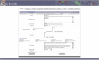
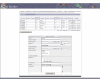
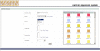
Results: BioBuilder is a Zope-based software tool that was developed to facilitate intuitive creation of protein databases. Protein data can be entered and annotated through web forms along with the flexibility to add customized annotation features to protein entries. A built-in review system permits a global team of scientists to coordinate their annotation efforts. We have already used BioBuilder to develop Human Protein Reference Database http://www.hprd.org, a comprehensive annotated repository of the human proteome. The data can be exported in the extensible markup language (XML) format, which is rapidly becoming as the standard format for data exchange.
Conclusions: As the proteomic data for several organisms begins to accumulate, BioBuilder will prove to be an invaluable platform for functional annotation and development of customizable protein centric databases. BioBuilder is open source and is available under the terms of LGPL.
Figures

References
-
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, Roechert B, Poux S, Jung E, Mersch H, Kersey P, Lappe M, Li Y, Zeng R, Rana D, Nikolski M, Husi H, Brun C, Shanker K, Grant SG, Sander C, Bork P, Zhu W, Pandey A, Brazma A, Jacq B, Vidal M, Sherman D, Legrain P, Cesareni G, Xenarios I, Eisenberg D, Steipe B, Hogue C, Apweiler R. The HUPO PSI Molecular Interaction Format – A community standard for the representation of protein interaction data. Nature Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. - DOI - PubMed
-
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of Human Protein Reference Database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. - DOI - PMC - PubMed
-
- Human Protein Reference Database http://www.hprd.org
-
- Zope Homepage http://www.zope.org
-
- Python Home page http://www.python.org
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

