Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression
- PMID: 15100397
- PMCID: PMC423217
- DOI: 10.1105/tpc.018986
Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression
Abstract
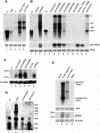
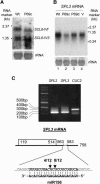
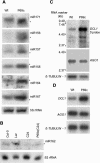
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are processed by the ribonuclease Dicer from distinct precursors, double-stranded RNA (dsRNA) and hairpin RNAs, respectively, although either may guide RNA silencing via a similar complex. The siRNA pathway is antiviral, whereas an emerging role for miRNAs is in the control of development. Here, we describe a virulence factor encoded by turnip yellow mosaic virus, p69, which suppresses the siRNA pathway but promotes the miRNA pathway in Arabidopsis thaliana. p69 suppression of the siRNA pathway is upstream of dsRNA and is as effective as genetic mutations in A. thaliana genes involved in dsRNA production. Possibly as a consequence of p69 suppression, p69-expressing plants contained elevated levels of a Dicer mRNA and of miRNAs as well as a correspondingly enhanced miRNA-guided cleavage of two host mRNAs. Because p69-expressing plants exhibited disease-like symptoms in the absence of viral infection, our findings suggest a novel mechanism for viral virulence by promoting the miRNA-guided inhibition of host gene expression.
Figures




Similar articles
-
Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways.Plant Cell. 2005 Nov;17(11):2873-85. doi: 10.1105/tpc.105.036608. Epub 2005 Oct 7. Plant Cell. 2005. PMID: 16214897 Free PMC article.
-
Fate of artificial microRNA-mediated resistance to plant viruses in mixed infections.Phytopathology. 2013 Aug;103(8):870-6. doi: 10.1094/PHYTO-09-12-0233-R. Phytopathology. 2013. PMID: 23617337
-
Artificial microRNA-mediated virus resistance in plants.J Virol. 2007 Jun;81(12):6690-9. doi: 10.1128/JVI.02457-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344304 Free PMC article.
-
Small but mighty RNA-mediated interference in plants.Indian J Exp Biol. 2005 Jan;43(1):7-24. Indian J Exp Biol. 2005. PMID: 15691061 Review.
-
Structural insights into the arms race between host and virus along RNA silencing pathways in Arabidopsis thaliana.Biol Rev Camb Philos Soc. 2014 May;89(2):337-55. doi: 10.1111/brv.12057. Epub 2013 Sep 4. Biol Rev Camb Philos Soc. 2014. PMID: 24034134 Review.
Cited by
-
Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems.Genome Biol. 2018 Sep 28;19(1):149. doi: 10.1186/s13059-018-1529-7. Genome Biol. 2018. PMID: 30266091 Free PMC article.
-
Silencing suppressors: viral weapons for countering host cell defenses.Protein Cell. 2011 Apr;2(4):273-81. doi: 10.1007/s13238-011-1037-y. Epub 2011 Apr 27. Protein Cell. 2011. PMID: 21528352 Free PMC article. Review.
-
RNA-binding protein immunoprecipitation as a tool to investigate plant miRNA processing interference by regulatory proteins of diverse origin.Plant Methods. 2018 Jan 31;14:9. doi: 10.1186/s13007-018-0276-9. eCollection 2018. Plant Methods. 2018. PMID: 29422942 Free PMC article.
-
ORF43 of maize rayado fino virus is dispensable for systemic infection of maize and transmission by leafhoppers.Virus Genes. 2016 Apr;52(2):303-7. doi: 10.1007/s11262-016-1287-0. Epub 2016 Feb 2. Virus Genes. 2016. PMID: 26837893
-
MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis.Cell. 2013 Apr 25;153(3):562-74. doi: 10.1016/j.cell.2013.04.005. Cell. 2013. PMID: 23622241 Free PMC article.
References
-
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279, 2113–2115. - PubMed
-
- Anandalakshmi, R., Marathe, R., Ge, X., Herr, J.M., Mau, C., Mallory, A., Pruss, G., Bowman, L., and Vance, V.B. (2000). A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290, 142–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

