EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ
- PMID: 15101985
- PMCID: PMC5517308
- DOI: 10.1111/j.1365-2958.2004.04016.x
EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ
Abstract
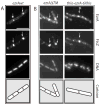
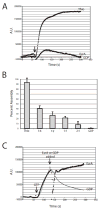
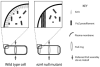
In response to a cell cycle signal, the cytoskeletal protein FtsZ assembles into a ring structure that establishes the location of the division site and serves as a framework for assembly of the division machinery. A battery of factors control FtsZ assembly to ensure that the ring forms in the correct position and at the precise time. EzrA, a negative regulator of FtsZ ring formation, is important for ensuring that the ring forms only once per cell cycle and that cytokinesis is restricted to mid-cell. EzrA is distributed throughout the plasma membrane and localizes to the ring in an FtsZ-dependent manner, suggesting that it interacts directly with FtsZ to modulate assembly. We have performed a series of experiments examining the interaction between EzrA and FtsZ. As little as twofold overexpression of EzrA blocks FtsZ ring formation in a sensitized genetic background, consistent with its predicted function. A purified EzrA fusion protein interacts directly with FtsZ to block assembly in vitro. Although EzrA is able to inhibit FtsZ assembly, it is unable to disassemble preformed polymers. These data support a model in which EzrA interacts directly with FtsZ at the plasma membrane to prevent polymerization and aberrant FtsZ ring formation.
Figures





Similar articles
-
A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ.Biochemistry. 2007 Sep 25;46(38):11013-22. doi: 10.1021/bi700710j. Epub 2007 Aug 24. Biochemistry. 2007. PMID: 17718511
-
The division inhibitor EzrA contains a seven-residue patch required for maintaining the dynamic nature of the medial FtsZ ring.J Bacteriol. 2007 Dec;189(24):9001-10. doi: 10.1128/JB.01172-07. Epub 2007 Sep 14. J Bacteriol. 2007. PMID: 17873055 Free PMC article.
-
Mechanism of regulation of prokaryotic tubulin-like GTPase FtsZ by membrane protein EzrA.J Biol Chem. 2007 May 18;282(20):14891-7. doi: 10.1074/jbc.M605177200. Epub 2006 Oct 16. J Biol Chem. 2007. PMID: 17043359
-
Strong FtsZ is with the force: mechanisms to constrict bacteria.Trends Microbiol. 2010 Aug;18(8):348-56. doi: 10.1016/j.tim.2010.06.001. Epub 2010 Jul 1. Trends Microbiol. 2010. PMID: 20598544 Review.
-
Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability.Annu Rev Microbiol. 2003;57:125-54. doi: 10.1146/annurev.micro.57.012903.074300. Annu Rev Microbiol. 2003. PMID: 14527275 Free PMC article. Review.
Cited by
-
A moonlighting enzyme links Escherichia coli cell size with central metabolism.PLoS Genet. 2013;9(7):e1003663. doi: 10.1371/journal.pgen.1003663. Epub 2013 Jul 25. PLoS Genet. 2013. PMID: 23935518 Free PMC article.
-
GpsB interacts with FtsZ in multiple species and may serve as an accessory Z-ring anchor.Mol Biol Cell. 2025 Jan 1;36(1):ar10. doi: 10.1091/mbc.E24-07-0302. Epub 2024 Nov 27. Mol Biol Cell. 2025. PMID: 39602291 Free PMC article.
-
Toward the Complete Functional Characterization of a Minimal Bacterial Proteome.J Phys Chem B. 2022 Sep 15;126(36):6820-6834. doi: 10.1021/acs.jpcb.2c04188. Epub 2022 Sep 1. J Phys Chem B. 2022. PMID: 36048731 Free PMC article.
-
Models versus pathogens: how conserved is the FtsZ in bacteria?Biosci Rep. 2023 Feb 27;43(2):BSR20221664. doi: 10.1042/BSR20221664. Biosci Rep. 2023. PMID: 36695643 Free PMC article. Review.
-
Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus.Appl Environ Microbiol. 2010 Jul;76(13):4346-53. doi: 10.1128/AEM.00359-10. Epub 2010 May 7. Appl Environ Microbiol. 2010. PMID: 20453129 Free PMC article.
References
-
- Addinall SG, Lutkenhaus J. FtsZ-spirals and –arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. - PubMed
-
- Arakawa T, Timasheff SN. Mechanism of protein salting in and salting out by divalent cation salts: balance between hydration and salt binding. Biochemistry. 1984;23:5912–5923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

