Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis
- PMID: 15101989
- PMCID: PMC1409746
- DOI: 10.1111/j.1365-2958.2004.04023.x
Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis
Abstract
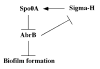
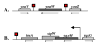
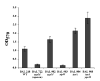
Bacillus subtilis is a ubiquitous soil bacterium that forms biofilms in a process that is negatively controlled by the transcription factor AbrB. To identify the AbrB-regulated genes required for biofilm formation by B. subtilis, genome-wide expression profiling studies of biofilms formed by spo0A abrB and sigH abrB mutant strains were performed. These data, in concert with previously published DNA microarray analysis of spo0A and sigH mutant strains, led to the identification of 39 operons that appear to be repressed by AbrB. Eight of these operons had previously been shown to be repressed by AbrB, and we confirmed AbrB repression for a further six operons by reverse transcription-PCR. The AbrB-repressed genes identified in this study are involved in processes known to be regulated by AbrB, such as extracellular degradative enzyme production and amino acid metabolism, and processes not previously known to be regulated by AbrB, such as membrane bioenergetics and cell wall functions. To determine whether any of these AbrB-regulated genes had a role in biofilm formation, we tested 23 mutants, each with a disruption in a different AbrB-regulated operon, for the ability to form biofilms. Two mutants had a greater than twofold defect in biofilm formation. A yoaW mutant exhibited a biofilm structure with reduced depth, and a sipW mutant exhibited only surface-attached cells and did not form a mature biofilm. YoaW is a putative secreted protein, and SipW is a signal peptidase. This is the first evidence that secreted proteins have a role in biofilm formation by Bacillus subtilis.
Figures

Similar articles
-
The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis.Mol Microbiol. 2001 Dec;42(5):1199-209. doi: 10.1046/j.1365-2958.2001.02709.x. Mol Microbiol. 2001. PMID: 11886552
-
SigmaX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh.J Bacteriol. 2009 Nov;191(22):6822-32. doi: 10.1128/JB.00618-09. Epub 2009 Sep 18. J Bacteriol. 2009. PMID: 19767430 Free PMC article.
-
DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis.J Bacteriol. 2009 Jan;191(1):100-8. doi: 10.1128/JB.01236-08. Epub 2008 Oct 31. J Bacteriol. 2009. PMID: 18978066 Free PMC article.
-
The Biofilm Regulatory Network from Bacillus subtilis: A Structure-Function Analysis.J Mol Biol. 2023 Feb 1;435(3):167923. doi: 10.1016/j.jmb.2022.167923. Epub 2022 Dec 16. J Mol Biol. 2023. PMID: 36535428 Review.
-
A complex path for domestication of B. subtilis sociality.Curr Genet. 2015 Nov;61(4):493-6. doi: 10.1007/s00294-015-0479-9. Epub 2015 Feb 14. Curr Genet. 2015. PMID: 25680358 Review.
Cited by
-
Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes.FEMS Microbiol Rev. 2015 Sep;39(5):649-69. doi: 10.1093/femsre/fuv015. Epub 2015 Apr 22. FEMS Microbiol Rev. 2015. PMID: 25907113 Free PMC article. Review.
-
Characterization of Bacillus subtilis Colony Biofilms via Mass Spectrometry and Fluorescence Imaging.J Proteome Res. 2016 Jun 3;15(6):1955-62. doi: 10.1021/acs.jproteome.6b00127. Epub 2016 May 11. J Proteome Res. 2016. PMID: 27136705 Free PMC article.
-
Cell death as a trigger for morphogenesis.PLoS One. 2018 Mar 22;13(3):e0191089. doi: 10.1371/journal.pone.0191089. eCollection 2018. PLoS One. 2018. PMID: 29565975 Free PMC article.
-
Influence of the sigmaB stress factor and yxaB, the gene for a putative exopolysaccharide synthase under sigmaB Control, on biofilm formation.J Bacteriol. 2008 May;190(10):3546-56. doi: 10.1128/JB.01665-07. Epub 2008 Mar 7. J Bacteriol. 2008. PMID: 18326573 Free PMC article.
-
New tools for comparing microscopy images: quantitative analysis of cell types in Bacillus subtilis.J Bacteriol. 2015 Feb 15;197(4):699-709. doi: 10.1128/JB.02501-14. Epub 2014 Dec 1. J Bacteriol. 2015. PMID: 25448819 Free PMC article.
References
-
- Cheggour A, Fanuel L, Duez C, Joris B, Bouillenne F, Devreese B, Van Driessche G, Van Beeumen J, Frere JM, Goffin C. The dppA gene of Bacillus subtilis encodes a new D-aminopeptidase. Mol Microbiol. 2000;38:504–513. - PubMed
-
- Cho HY, Kim YW, Kim TJ, Lee HS, Kim DY, Kim JW, Lee YW, Leed S, Park KH. Molecular characterization of a dimeric intracellular maltogenic amylase of Bacillus subtilis SUH4-2. Biochim Biophys Acta. 2000;1478:333–340. - PubMed
-
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

