Catabolite activator protein: DNA binding and transcription activation
- PMID: 15102444
- PMCID: PMC2765107
- DOI: 10.1016/j.sbi.2004.01.012
Catabolite activator protein: DNA binding and transcription activation
Abstract
Recently determined structures of the Escherichia coli catabolite activator protein (CAP) in complex with DNA, and in complex with the RNA polymerase alpha subunit C-terminal domain (alphaCTD) and DNA, have yielded insights into how CAP binds DNA and activates transcription. Comparison of multiple structures of CAP-DNA complexes has revealed the contributions of direct and indirect readout to DNA binding by CAP. The structure of the CAP-alphaCTD-DNA complex has provided the first structural description of interactions between a transcription activator and its functional target within the general transcription machinery. Using the structure of the CAP-alphaCTD-DNA complex, the structure of an RNA polymerase-DNA complex, and restraints from biophysical, biochemical and genetic experiments, it has been possible to construct detailed three-dimensional models of intact class I and class II transcription activation complexes.
Figures
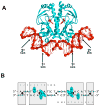
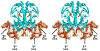
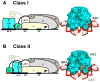


References
-
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. - PubMed
-
- Rhodius VA, Busby SJ. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. - PubMed
-
- Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. - PubMed
-
- Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304:847–859. - PubMed
-
- Chu SY, Tordova M, Gilliland GL, Gorshkova I, Shi Y, Wang S, Schwarz FP. The structure of the T127L/S128A mutant of cAMP receptor protein facilitates promoter site binding. J Biol Chem. 2001;276:11230–11236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

