Analysis of haptoglobin and hemoglobin-haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry
- PMID: 15102756
- PMCID: PMC387877
- DOI: 10.1128/IAI.72.5.2494-2506.2004
Analysis of haptoglobin and hemoglobin-haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry
Abstract
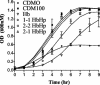
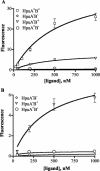
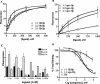
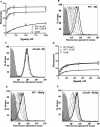
Neisseria meningitidis expresses a two-component TonB-dependent receptor, HpuAB, which mediates heme-iron (Hm-Fe) acquisition from hemoglobin and hemoglobin-haptoglobin complexes. Due to genetic polymorphisms in the human haptoglobin gene, haptoglobin (and hemoglobin-haptoglobin) exists as three structurally distinct phenotypes. In this study, we examined the influence of the haptoglobin phenotype on the interactions of HpuAB with apo-haptoglobin and hemoglobin-haptoglobin. Growth assays confirmed that HpuAB utilizes hemoglobin-haptoglobin more efficiently than hemoglobin as an Fe source and revealed a preference for human-specific, polymeric 2-2 or 2-1 hemoglobin-haptoglobin complexes. We developed a flow cytometry-based assay to measure the binding kinetics of fluorescein-labeled ligands to HpuAB on live, intact meningococci. The binding affinity of HpuAB for hemoglobin-haptoglobin phenotypes correlated well with the ability of each ligand to support Neisseria meningitidis growth, with higher affinities exhibited for types 2-2 and 2-1 hemoglobin-haptoglobin. Saturable binding of Hb and apo-haptoglobin suggested that HpuAB-mediated utilization of hemoglobin-haptoglobin involves specific interactions with both components. In contrast to previous studies, we detected binding of HpuB expressed alone to hemoglobin, apo-haptoglobin, and hemoglobin-haptoglobin of all three phenotypes. However, in the absence of HpuA, the binding capacity and/or affinity of the receptor was reduced and the dissociation of hemoglobin was impaired. We did not detect binding of HpuA alone to hemoglobin, apo-haptoglobin, or hemoglobin-haptoglobin; however, the lipoprotein is crucial for optimal recognition and use of ligands by the receptor. Finally, this study confirmed the integral role of TonB and the proton motive force in the binding and dissociation of Hb and hemoglobin-haptoglobin from HpuAB.
Figures
Similar articles
-
Interactions of haemoglobin with the Neisseria meningitidis receptor HpuAB: the role of TonB and an intact proton motive force.Mol Microbiol. 2002 Jan;43(2):335-54. doi: 10.1046/j.1365-2958.2002.02745.x. Mol Microbiol. 2002. PMID: 11985713
-
Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2.Mol Microbiol. 1999 Jun;32(5):977-89. doi: 10.1046/j.1365-2958.1999.01409.x. Mol Microbiol. 1999. PMID: 10361300
-
Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis.Mol Microbiol. 1997 Feb;23(4):737-49. doi: 10.1046/j.1365-2958.1997.2501619.x. Mol Microbiol. 1997. PMID: 9157245
-
Iron transport systems in Neisseria meningitidis.Microbiol Mol Biol Rev. 2004 Mar;68(1):154-71. doi: 10.1128/MMBR.68.1.154-171.2004. Microbiol Mol Biol Rev. 2004. PMID: 15007100 Free PMC article. Review.
-
Mechanisms of iron acquisition by the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae.Front Biosci. 2003 Sep 1;8:d1186-218. doi: 10.2741/1133. Front Biosci. 2003. PMID: 12957813 Review.
Cited by
-
Extracellular heme uptake and the challenges of bacterial cell membranes.Curr Top Membr. 2012;69:359-92. doi: 10.1016/B978-0-12-394390-3.00013-6. Curr Top Membr. 2012. PMID: 23046657 Free PMC article.
-
The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.Proc Natl Acad Sci U S A. 2023 Jan 31;120(5):e2211939120. doi: 10.1073/pnas.2211939120. Epub 2023 Jan 24. Proc Natl Acad Sci U S A. 2023. PMID: 36693107 Free PMC article.
-
Kinetic analysis of ligand interaction with the gonococcal transferrin-iron acquisition system.Biometals. 2009 Jun;22(3):439-51. doi: 10.1007/s10534-008-9179-y. Epub 2008 Dec 2. Biometals. 2009. PMID: 19048191 Free PMC article.
-
An Exposed Outer Membrane Hemin-Binding Protein Facilitates Hemin Transport by a TonB-Dependent Receptor in Riemerella anatipestifer.Appl Environ Microbiol. 2021 Jul 13;87(15):e0036721. doi: 10.1128/AEM.00367-21. Epub 2021 Jul 13. Appl Environ Microbiol. 2021. PMID: 33990314 Free PMC article.
-
Host iron binding proteins acting as niche indicators for Neisseria meningitidis.PLoS One. 2009;4(4):e5198. doi: 10.1371/journal.pone.0005198. Epub 2009 Apr 8. PLoS One. 2009. PMID: 19352437 Free PMC article.
References
-
- Braun, V., S. Gaisser, C. Herrmann, K. Kampfenkel, H. Killmann, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

