Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity
- PMID: 15102773
- PMCID: PMC387885
- DOI: 10.1128/IAI.72.5.2648-2658.2004
Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity
Abstract
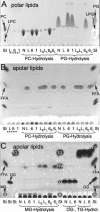
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular pathogen of amoebae, macrophages, and epithelial cells. The pathology of Legionella infections involves alveolar cell destruction, and several proteins of L. pneumophila are known to contribute to this ability. By screening a genomic library of L. pneumophila, we found an additional L. pneumophila gene, plaB, which coded for a hemolytic activity and contained a lipase consensus motif in its deduced protein sequence. Moreover, Escherichia coli harboring the L. pneumophila plaB gene showed increased activity in releasing fatty acids predominantly from diacylphospho- and lysophospholipids, demonstrating that it encodes a phospholipase A. It has been reported that culture supernatants and cell lysates of L. pneumophila possess phospholipase A activity; however, only the major secreted lysophospholipase A PlaA has been investigated on the molecular level. We therefore generated isogenic L. pneumophila plaB mutants and tested those for hemolysis, lipolytic activities, and intracellular survival in amoebae and macrophages. Compared to wild-type L. pneumophila, the plaB mutant showed reduced hemolysis of human red blood cells and almost completely lost its cell-associated lipolytic activity. We conclude that L. pneumophila plaB is the gene encoding the major cell-associated phospholipase A, possibly contributing to bacterial cytotoxicity due to its hemolytic activity. On the other hand, in view of the fact that the plaB mutant multiplied like the wild type both in U937 macrophages and in Acanthamoeba castellanii amoebae, plaB is not essential for intracellular survival of the pathogen.
Figures





Similar articles
-
Phospholipase PlaB of Legionella pneumophila represents a novel lipase family: protein residues essential for lipolytic activity, substrate specificity, and hemolysis.J Biol Chem. 2009 Oct 2;284(40):27185-94. doi: 10.1074/jbc.M109.026021. Epub 2009 Jul 29. J Biol Chem. 2009. PMID: 19640837 Free PMC article.
-
Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine.Infect Immun. 2002 Nov;70(11):6094-106. doi: 10.1128/IAI.70.11.6094-6106.2002. Infect Immun. 2002. PMID: 12379686 Free PMC article.
-
Characterization of the major secreted zinc metalloprotease- dependent glycerophospholipid:cholesterol acyltransferase, PlaC, of Legionella pneumophila.Infect Immun. 2005 May;73(5):2899-909. doi: 10.1128/IAI.73.5.2899-2909.2005. Infect Immun. 2005. PMID: 15845496 Free PMC article.
-
The manifold phospholipases A of Legionella pneumophila - identification, export, regulation, and their link to bacterial virulence.Int J Med Microbiol. 2008 Apr;298(3-4):169-81. doi: 10.1016/j.ijmm.2007.11.004. Epub 2008 Jan 4. Int J Med Microbiol. 2008. PMID: 18178130 Review.
-
Characterisation of Legionella pneumophila phospholipases and their impact on host cells.Eur J Cell Biol. 2011 Nov;90(11):903-12. doi: 10.1016/j.ejcb.2010.12.003. Epub 2011 Feb 20. Eur J Cell Biol. 2011. PMID: 21342713 Review.
Cited by
-
Identification and characterization of hemolysin-like proteins similar to RTX toxin in Pasteurella pneumotropica.J Bacteriol. 2009 Jun;191(11):3698-705. doi: 10.1128/JB.01527-08. Epub 2009 Apr 10. J Bacteriol. 2009. PMID: 19363112 Free PMC article.
-
Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome.Prog Lipid Res. 2010 Jan;49(1):1-26. doi: 10.1016/j.plipres.2009.07.003. Epub 2009 Jul 26. Prog Lipid Res. 2010. PMID: 19638285 Free PMC article. Review.
-
Molecular pathogenesis of infections caused by Legionella pneumophila.Clin Microbiol Rev. 2010 Apr;23(2):274-98. doi: 10.1128/CMR.00052-09. Clin Microbiol Rev. 2010. PMID: 20375353 Free PMC article. Review.
-
Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles.Infect Immun. 2008 May;76(5):1825-36. doi: 10.1128/IAI.01396-07. Epub 2008 Feb 4. Infect Immun. 2008. PMID: 18250176 Free PMC article.
-
Comparative analysis of Legionella lytica genome identifies specific metabolic traits and virulence factors.Sci Rep. 2025 Feb 14;15(1):5554. doi: 10.1038/s41598-025-90154-5. Sci Rep. 2025. PMID: 39952999 Free PMC article.
References
-
- Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

