Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles
- PMID: 15102912
- PMCID: PMC6729412
- DOI: 10.1523/JNEUROSCI.5259-03.2004
Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles
Abstract
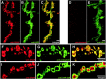
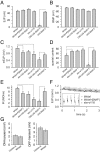
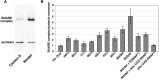
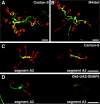
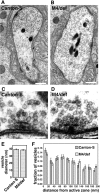
The N-ethylmaleimide-sensitive factor (NSF) and soluble NSF attachment protein (SNAP) are cytosolic factors that promote vesicle fusion with a target membrane in both the constitutive and regulated secretory pathways. NSF and SNAP are thought to function by catalyzing the disassembly of a SNAP receptor (SNARE) complex consisting of membrane proteins of the secretory vesicle and target membrane. Although studies of NSF function have provided strong support for this model, the precise biochemical role of SNAP remains controversial. To further explore the function of SNAP, we have used mutational and transgenic approaches in Drosophila to investigate the effect of altered SNAP dosage on neurotransmitter release and SNARE complex metabolism. Our results indicate that reduced SNAP activity results in diminished neurotransmitter release and accumulation of a neural SNARE complex. Increased SNAP dosage results in defective synapse formation and a variety of tissue morphological defects without detectably altering the abundance of neural SNARE complexes. The SNAP overexpression phenotypes are enhanced by mutations in other secretory components and are at least partially overcome by co-overexpression of NSF, suggesting that these phenotypes derive from a specific perturbation of the secretory pathway. Our results indicate that SNAP promotes neurotransmitter release and SNARE complex disassembly but inhibits secretion when present at high abundance relative to NSF.
Figures

References
-
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS (2002) Wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila Neuron 33: 545-558. - PubMed
-
- Bellen HJ, Budnik V (2000) The neuromuscular junction. In: Drosophila protocols (Sullivan W, Hawley RS, Ashburner M, eds), pp 175-199. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
