Genetic and molecular characterization of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae with unusually high resistance to ampicillin
- PMID: 15105114
- PMCID: PMC400547
- DOI: 10.1128/AAC.48.5.1630-1639.2004
Genetic and molecular characterization of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae with unusually high resistance to ampicillin
Abstract
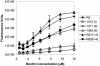
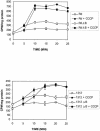
Previous studies with beta-lactamase-negative, ampicillin-resistant (BLNAR) Haemophilus influenzae from Japan, France, and North America indicate that mutations in ftsI encoding PBP3 confer ampicillin MICs of 1 to 4 micro g/ml. Several BLNAR strains with ampicillin MICs of 4 to 16 micro g/ml recently isolated from North America were studied. Pulsed-field gel electrophoresis identified 12 unique BLNAR strains; sequencing of their ftsI transpeptidase domains identified 1 group I and 11 group II mutants, as designated previously (K. Ubukata, Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno, Antimicrob. Agents Chemother. 45:1693-1699, 2001). Geometric mean ampicillin MICs for several clinical isolates were 8 to 10.56 micro g/ml. Replacement of the ftsI gene in H. influenzae Rd with the intact ftsI from several clinical isolates resulted in integrants with typical BLNAR geometric mean ampicillin MICs of 1.7 to 2.2 micro g/ml. Cloning and purification of His-tagged PBP3 from three clinical BLNAR strains showed significantly reduced Bocillin binding compared to that of PBP3 from strain Rd. Based on these data, changes in PBP3 alone could not account for the high ampicillin MICs observed for these BLNAR isolates. In an effort to determine the presence of additional mechanism(s) of ampicillin resistance, sequencing of the transpeptidase regions of pbp1a, -1b, and -2 was performed. While numerous changes were observed compared to the sequences from Rd, no consistent pattern correlating with high-level ampicillin resistance was apparent. Additional analysis of the resistant BLNAR strains revealed frame shift insertions in acrR for all four high-level, ampicillin-resistant isolates. acrR was intact for all eight low-level ampicillin-resistant and four ampicillin-susceptible strains tested. A knockout of acrB made in one clinical isolate (initial mean ampicillin MIC of 10.3 micro g/ml) lowered the ampicillin MIC to 3.67 micro g/ml, typical for BLNAR strains. These studies illustrate that BLNAR strains with high ampicillin MICs exist that have combined resistance mechanisms in PBP3 and in the AcrAB efflux pump.
Figures

Similar articles
-
Rapidly increasing prevalence of beta-lactamase-nonproducing, ampicillin-resistant Haemophilus influenzae type b in patients with meningitis.Antimicrob Agents Chemother. 2004 May;48(5):1509-14. doi: 10.1128/AAC.48.5.1509-1514.2004. Antimicrob Agents Chemother. 2004. PMID: 15105098 Free PMC article.
-
Contribution of beta-lactamase and PBP amino acid substitutions to amoxicillin/clavulanate resistance in beta-lactamase-positive, amoxicillin/clavulanate-resistant Haemophilus influenzae.J Antimicrob Chemother. 2003 Dec;52(6):1018-21. doi: 10.1093/jac/dkg474. Epub 2003 Oct 29. J Antimicrob Chemother. 2003. PMID: 14585854
-
Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae.Antimicrob Agents Chemother. 2001 Jun;45(6):1693-9. doi: 10.1128/AAC.45.6.1693-1699.2001. Antimicrob Agents Chemother. 2001. PMID: 11353613 Free PMC article.
-
[Beta-lactamase negative ampicillin-resistant Haemophilus influenzae(BLNAR)].Nihon Rinsho. 2001 Apr;59(4):688-93. Nihon Rinsho. 2001. PMID: 11304990 Review. Japanese.
-
[BLNAR (beta-lactamase-nonproducing ampicillin-resistant Haemophilus influenzae)].Nihon Rinsho. 2003 Mar;61 Suppl 3:176-81. Nihon Rinsho. 2003. PMID: 12717968 Review. Japanese. No abstract available.
Cited by
-
Heterologous viral expression systems in fosmid vectors increase the functional analysis potential of metagenomic libraries.Sci Rep. 2013;3:1107. doi: 10.1038/srep01107. Epub 2013 Jan 22. Sci Rep. 2013. PMID: 23346364 Free PMC article.
-
Three decades of beta-lactamase inhibitors.Clin Microbiol Rev. 2010 Jan;23(1):160-201. doi: 10.1128/CMR.00037-09. Clin Microbiol Rev. 2010. PMID: 20065329 Free PMC article. Review.
-
Polymorphism in ftsI gene and {beta}-lactam susceptibility in Portuguese Haemophilus influenzae strains: clonal dissemination of beta-lactamase-positive isolates with decreased susceptibility to amoxicillin/clavulanic acid.J Antimicrob Chemother. 2011 Apr;66(4):788-96. doi: 10.1093/jac/dkq533. Epub 2011 Jan 25. J Antimicrob Chemother. 2011. PMID: 21393206 Free PMC article.
-
Yersinia pestis AcrAB-TolC in antibiotic resistance and virulence.Antimicrob Agents Chemother. 2012 Feb;56(2):1120-3. doi: 10.1128/AAC.05338-11. Epub 2011 Nov 14. Antimicrob Agents Chemother. 2012. PMID: 22083483 Free PMC article.
-
Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria.Clin Microbiol Rev. 2006 Apr;19(2):382-402. doi: 10.1128/CMR.19.2.382-402.2006. Clin Microbiol Rev. 2006. PMID: 16614254 Free PMC article. Review.
References
-
- Arbing, M. A., J. W. Hanrahan, and J. W. Coulton. 2002. Altered channel properties of porins from Haemophilus influenzae: isolates from cystic fibrosis patients. J. Membr. Biol. 189:131-141. - PubMed
-
- Barcak, G. J., M. S. Chandler, R. J. Redfield, and J.-F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. - PubMed
-
- Burns, J. L., and A. L. Smith. 1987. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J. Gen. Microbiol. 133:1273-1277. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

