Biochemical characterization of Streptococcus pneumoniae penicillin-binding protein 2b and its implication in beta-lactam resistance
- PMID: 15105143
- PMCID: PMC400559
- DOI: 10.1128/AAC.48.5.1848-1855.2004
Biochemical characterization of Streptococcus pneumoniae penicillin-binding protein 2b and its implication in beta-lactam resistance
Abstract
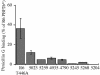
Extensive use of beta-lactam antibiotics has led to the selection of pathogenic streptococci resistant to beta-lactams due to modifications of the penicillin-binding proteins (PBPs). PBP2b from Streptococcus pneumoniae is a monofunctional (class B) high-molecular-weight PBP catalyzing the transpeptidation between adjacent stem peptides of peptidoglycan. The transpeptidase domain of PBP2b isolated from seven clinical resistant (CR) strains contains 7 to 44 amino acid changes over the sequence of PBP2b from the R6 beta-lactam-sensitive strain. We show that the extracellular soluble domains of recombinant PBP2b proteins (PBP2b*) originating from these CR strains have an in vitro affinity for penicillin G that is reduced by up to 99% from that of the R6 strain. The Thr446Ala mutation is always observed in CR strains and is close to the key conserved motif (S(443)SN). The Thr446Ala mutation in R6 PBP2b* displays a 60% reduction in penicillin G affinity in vitro compared to that for the wild-type protein. A recombinant R6 strain expressing the R6 PBP2b Thr446Ala mutation is twofold less sensitive to piperacillin than the parental S. pneumoniae strain. Analysis of the Thr446Ala mutation in the context of the PBP2b CR sequences revealed that its influence depends upon the presence of other unidentified mutations.
Figures
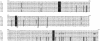

References
-
- Chesnel, L., L. Pernot, D. Lemaire, D. Champelovier, J. Croize, O. Dideberg, T. Vernet, and A. Zapun. 2003. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J. Biol. Chem. 278:44448-44456. - PubMed
-
- Di Guilmi, A. M., N. Mouz, J. P. Andrieu, J. Hoskins, S. R. Jaskunas, J. Gagnon, O. Dideberg, and T. Vernet. 1998. Identification, purification, and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniae penicillin-binding protein 1a. J. Bacteriol. 180:5652-5659. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

