RISC is a 5' phosphomonoester-producing RNA endonuclease
- PMID: 15105377
- PMCID: PMC406288
- DOI: 10.1101/gad.1187904
RISC is a 5' phosphomonoester-producing RNA endonuclease
Abstract
Gene silencing in the process of RNA interference is mediated by a ribonucleoprotein complex referred to as RNA-induced silencing complex (RISC). Here we describe the molecular mechanism of target RNA cleavage using affinity-purified minimal RISC from human cells. Cleavage proceeds via hydrolysis and the release of a 3'-hydroxyl and a 5'-phosphate terminus. Substitution of the 2'-hydroxyl group at the cleavage site by 2'-deoxy had no significant effect, suggesting that product release and/or a conformational transition rather than a chemical step is rate-limiting. Substitution by 2'-O-methyl at the cleavage site substantially reduced cleavage, which is presumably due to steric interference. Mutational analysis of the target RNA revealed that mismatches across from the 5' or the 3' end of the siRNA had little effect and that substrate RNAs as short as 15 nucleotides were cleaved by RISC.
Figures
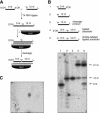
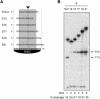

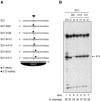
Similar articles
-
Molecular basis for target RNA recognition and cleavage by human RISC.Cell. 2007 Jul 13;130(1):101-12. doi: 10.1016/j.cell.2007.04.037. Cell. 2007. PMID: 17632058
-
Cleavage of the siRNA passenger strand during RISC assembly in human cells.EMBO Rep. 2006 Mar;7(3):314-20. doi: 10.1038/sj.embor.7400637. Epub 2006 Jan 20. EMBO Rep. 2006. PMID: 16439995 Free PMC article.
-
The RNA-induced silencing complex is a Mg2+-dependent endonuclease.Curr Biol. 2004 May 4;14(9):787-91. doi: 10.1016/j.cub.2004.03.008. Curr Biol. 2004. PMID: 15120070
-
Life of RISC: Formation, action, and degradation of RNA-induced silencing complex.Mol Cell. 2022 Jan 6;82(1):30-43. doi: 10.1016/j.molcel.2021.11.026. Epub 2021 Dec 22. Mol Cell. 2022. PMID: 34942118 Review.
-
RISC assembly: Coordination between small RNAs and Argonaute proteins.Biochim Biophys Acta. 2016 Jan;1859(1):71-81. doi: 10.1016/j.bbagrm.2015.08.007. Epub 2015 Aug 22. Biochim Biophys Acta. 2016. PMID: 26303205 Review.
Cited by
-
Overview of microRNA biology.Semin Liver Dis. 2015 Feb;35(1):3-11. doi: 10.1055/s-0034-1397344. Epub 2015 Jan 29. Semin Liver Dis. 2015. PMID: 25632930 Free PMC article. Review.
-
MicroRNA-200a regulates Grb2 and suppresses differentiation of mouse embryonic stem cells into endoderm and mesoderm.PLoS One. 2013 Jul 18;8(7):e68990. doi: 10.1371/journal.pone.0068990. Print 2013. PLoS One. 2013. PMID: 23874841 Free PMC article.
-
Rapid and specific purification of Argonaute-small RNA complexes from crude cell lysates.RNA. 2013 Feb;19(2):271-9. doi: 10.1261/rna.036921.112. Epub 2012 Dec 18. RNA. 2013. PMID: 23249751 Free PMC article.
-
MicroRNA in carcinogenesis & cancer diagnostics: a new paradigm.Indian J Med Res. 2013 Apr;137(4):680-94. Indian J Med Res. 2013. PMID: 23703335 Free PMC article. Review.
-
Active turnover modulates mature microRNA activity in Caenorhabditis elegans.Nature. 2009 Sep 24;461(7263):546-9. doi: 10.1038/nature08349. Epub 2009 Sep 6. Nature. 2009. PMID: 19734881
References
-
- Alexander M., Heppel, L.A., and Hurwitz, J. 1961. The purification and properties of micrococcal nuclease. J. Biol. Chem. 236: 3014-3019. - PubMed
-
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. - PubMed
-
- Braasch D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., and Corey, D.R. 2003. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42: 7967-7975. - PubMed
-
- Capodici J., Kariko, K., and Weissman, D. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169: 5196-5201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
