Folate biosynthesis in higher plants. cDNA cloning, heterologous expression, and characterization of dihydroneopterin aldolases
- PMID: 15107504
- PMCID: PMC429337
- DOI: 10.1104/pp.103.038430
Folate biosynthesis in higher plants. cDNA cloning, heterologous expression, and characterization of dihydroneopterin aldolases
Abstract
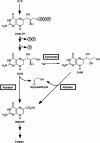
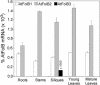
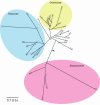
Dihydroneopterin aldolase (EC 4.1.2.25) is one of the enzymes of folate synthesis that remains to be cloned and characterized from plants. This enzyme catalyzes conversion of 7,8-dihydroneopterin (DHN) to 6-hydroxymethyl-7,8-dihydropterin, and is encoded by the folB gene in Escherichia coli. The E. coli FolB protein also mediates epimerization of DHN to 7,8-dihydromonapterin. Searches of the Arabidopsis genome detected three genes encoding substantially diverged FolB homologs (AtFolB1-3, sharing 57%-73% identity), for which cDNAs were isolated. A fourth cDNA specifying a FolB-like protein (LeFolB1) was obtained from tomato (Lycopersicon esculentum) by reverse transcription-PCR. When overproduced in E. coli, recombinant AtFolB1, AtFolB2, and LeFolB1 proteins all had both dihydroneopterin aldolase and epimerase activities, and carried out the aldol cleavage reaction on the epimerization product, 7,8-dihydromonapterin, as well as on DHN. AtFolB3, however, could not be expressed in active form. Size exclusion chromatography indicated that the plant enzyme is an octamer, like the bacterial enzyme. Quantifying expression of the Arabidopsis genes by real-time reverse transcription-PCR showed that AtFolB1 and AtFolB2 messages occur at low levels throughout the plant, whereas the AtFolB3 mRNA was detected only in siliques and only with an extremely low abundance. Sequence comparisons and phylogenetic analysis of FolB homologs from 16 plants indicated that their N-terminal regions are highly variable, and that most species have a small number of FolB genes that diverged after separation of the lineages leading to families. The substantial divergence of FolB homologs in Arabidopsis and other plants suggests that some of them may act on substrates other than DHN.
Figures

Similar articles
-
Biosynthesis of pteridines in Escherichia coli. Structural and mechanistic similarity of dihydroneopterin-triphosphate epimerase and dihydroneopterin aldolase.J Biol Chem. 1998 Jul 10;273(28):17418-24. doi: 10.1074/jbc.273.28.17418. J Biol Chem. 1998. PMID: 9651328
-
6-pyruvoyltetrahydropterin synthase paralogs replace the folate synthesis enzyme dihydroneopterin aldolase in diverse bacteria.J Bacteriol. 2009 Jul;191(13):4158-65. doi: 10.1128/JB.00416-09. Epub 2009 Apr 24. J Bacteriol. 2009. PMID: 19395485 Free PMC article.
-
Folate synthesis in plants: the first step of the pterin branch is mediated by a unique bimodular GTP cyclohydrolase I.Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12489-94. doi: 10.1073/pnas.192278499. Epub 2002 Sep 9. Proc Natl Acad Sci U S A. 2002. PMID: 12221287 Free PMC article.
-
Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme.J Mol Biol. 2005 May 27;349(1):61-72. doi: 10.1016/j.jmb.2005.03.023. Epub 2005 Apr 2. J Mol Biol. 2005. PMID: 15876368
-
Beta galactosidases in Arabidopsis and tomato - a mini review.Biochem Soc Trans. 2016 Feb;44(1):150-8. doi: 10.1042/BST20150217. Biochem Soc Trans. 2016. PMID: 26862200 Review.
Cited by
-
Antioxidants in Potatoes: A Functional View on One of the Major Food Crops Worldwide.Molecules. 2021 Apr 22;26(9):2446. doi: 10.3390/molecules26092446. Molecules. 2021. PMID: 33922183 Free PMC article. Review.
-
Genetic and physiological regulation of folate in pak choi (Brassica rapa subsp. Chinensis) germplasm.J Exp Bot. 2020 Aug 6;71(16):4914-4929. doi: 10.1093/jxb/eraa218. J Exp Bot. 2020. PMID: 32639001 Free PMC article.
-
Genome-wide identification and transcriptional analysis of folate metabolism-related genes in maize kernels.BMC Plant Biol. 2015 Aug 19;15:204. doi: 10.1186/s12870-015-0578-2. BMC Plant Biol. 2015. PMID: 26283542 Free PMC article.
-
Evolution of folate biosynthesis and metabolism across algae and land plant lineages.Sci Rep. 2019 Apr 5;9(1):5731. doi: 10.1038/s41598-019-42146-5. Sci Rep. 2019. PMID: 30952916 Free PMC article.
-
Folate biofortification in tomatoes by engineering the pteridine branch of folate synthesis.Proc Natl Acad Sci U S A. 2004 Sep 21;101(38):13720-5. doi: 10.1073/pnas.0404208101. Epub 2004 Sep 13. Proc Natl Acad Sci U S A. 2004. PMID: 15365185 Free PMC article.
References
-
- Basset GJC, Quinlivan EP, Ravanel S, Rébeillé F, Nichols BP, Shinozaki K, Seki M, Adams-Phillips LC, Giovannoni JJ, Gregory JF III (2004) Folate synthesis in plants: the p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids. Proc Natl Acad Sci USA 101: 1496–1501 - PMC - PubMed
-
- Bermingham A, Derrick JP (2002) The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24: 637–648 - PubMed
-
- Bouis HE (2002) Plant breeding: a new tool for fighting micronutrient malnutrition. J Nutr 132: 491S–494S - PubMed
-
- Bullock WO, Fernandez JM, Short JM (1987) XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5: 376–379
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

