The G protein-coupled receptor rhodopsin in the native membrane
- PMID: 15111110
- PMCID: PMC1393389
- DOI: 10.1016/S0014-5793(04)00194-2
The G protein-coupled receptor rhodopsin in the native membrane
Abstract
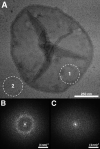
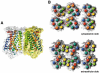
The higher-order structure of G protein-coupled receptors (GPCRs) in membranes may involve dimerization and formation of even larger oligomeric complexes. Here, we have investigated the organization of the prototypical GPCR rhodopsin in its native membrane by electron and atomic force microscopy (AFM). Disc membranes from mice were isolated and observed by AFM at room temperature. In all experimental conditions, rhodopsin forms structural dimers organized in paracrystalline arrays. A semi-empirical molecular model for the rhodopsin paracrystal is presented validating our previously reported results. Finally, we compare our model with other currently available models describing the supramolecular structure of GPCRs in the membrane.
Figures


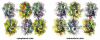
Similar articles
-
The supramolecular structure of the GPCR rhodopsin in solution and native disc membranes.Mol Membr Biol. 2004 Nov-Dec;21(6):435-46. doi: 10.1080/09687860400020291. Mol Membr Biol. 2004. PMID: 15764373 Free PMC article.
-
Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors.Curr Opin Struct Biol. 2006 Apr;16(2):252-9. doi: 10.1016/j.sbi.2006.03.013. Epub 2006 Mar 29. Curr Opin Struct Biol. 2006. PMID: 16567090 Review.
-
Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes.J Biol Chem. 2003 Jun 13;278(24):21655-21662. doi: 10.1074/jbc.M302536200. Epub 2003 Mar 27. J Biol Chem. 2003. PMID: 12663652 Free PMC article.
-
G protein-coupled receptor rhodopsin.Annu Rev Biochem. 2006;75:743-67. doi: 10.1146/annurev.biochem.75.103004.142743. Annu Rev Biochem. 2006. PMID: 16756510 Free PMC article. Review.
-
Atomic-force microscopy: Rhodopsin dimers in native disc membranes.Nature. 2003 Jan 9;421(6919):127-8. doi: 10.1038/421127a. Nature. 2003. PMID: 12520290 No abstract available.
Cited by
-
Membrane protein assembly into Nanodiscs.FEBS Lett. 2010 May 3;584(9):1721-7. doi: 10.1016/j.febslet.2009.10.024. Epub 2009 Oct 16. FEBS Lett. 2010. PMID: 19836392 Free PMC article. Review.
-
Unfolding and extraction of a transmembrane alpha-helical peptide: dynamic force spectroscopy and molecular dynamics simulations.Biophys J. 2005 Nov;89(5):3129-40. doi: 10.1529/biophysj.105.061721. Epub 2005 Aug 5. Biophys J. 2005. PMID: 16085762 Free PMC article.
-
Differential distribution of proteins and lipids in detergent-resistant and detergent-soluble domains in rod outer segment plasma membranes and disks.J Neurochem. 2008 Jan;104(2):336-52. doi: 10.1111/j.1471-4159.2007.04971.x. Epub 2007 Oct 18. J Neurochem. 2008. PMID: 17944869 Free PMC article.
-
A day in the life of a G protein-coupled receptor: the contribution to function of G protein-coupled receptor dimerization.Br J Pharmacol. 2008 Mar;153 Suppl 1(Suppl 1):S216-29. doi: 10.1038/sj.bjp.0707490. Epub 2007 Oct 29. Br J Pharmacol. 2008. PMID: 17965750 Free PMC article. Review.
-
Vertebrate membrane proteins: structure, function, and insights from biophysical approaches.Pharmacol Rev. 2008 Mar;60(1):43-78. doi: 10.1124/pr.107.07111. Epub 2008 Mar 5. Pharmacol Rev. 2008. PMID: 18321962 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

