Comparing gene discovery from Affymetrix GeneChip microarrays and Clontech PCR-select cDNA subtraction: a case study
- PMID: 15113399
- PMCID: PMC415544
- DOI: 10.1186/1471-2164-5-26
Comparing gene discovery from Affymetrix GeneChip microarrays and Clontech PCR-select cDNA subtraction: a case study
Abstract
Background: Several high throughput technologies have been employed to identify differentially regulated genes that may be molecular targets for drug discovery. Here we compared the sets of differentially regulated genes discovered using two experimental approaches: a subtracted suppressive hybridization (SSH) cDNA library methodology and Affymetrix GeneChip technology. In this "case study" we explored the transcriptional pattern changes during the in vitro differentiation of human monocytes to myeloid dendritic cells (DC), and evaluated the potential for novel gene discovery using the SSH methodology.
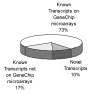
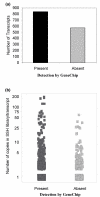
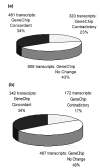
Results: The same RNA samples isolated from peripheral blood monocyte precursors and immature DC (iDC) were used for GeneChip microarray probing and SSH cDNA library construction. 10,000 clones from each of the two-way SSH libraries (iDC-monocytes and monocytes-iDC) were picked for sequencing. About 2000 transcripts were identified for each library from 8000 successful sequences. Only 70% to 75% of these transcripts were represented on the U95 series GeneChip microarrays, implying that 25% to 30% of these transcripts might not have been identified in a study based only on GeneChip microarrays. In addition, about 10% of these transcripts appeared to be "novel", although these have not yet been closely examined. Among the transcripts that are also represented on the chips, about a third were concordantly discovered as differentially regulated between iDC and monocytes by GeneChip microarray transcript profiling. The remaining two thirds were either not inferred as differentially regulated from GeneChip microarray data, or were called differentially regulated but in the opposite direction. This underscores the importance both of generating reciprocal pairs of SSH libraries, and of real-time RT-PCR confirmation of the results.
Conclusions: This study suggests that SSH could be used as an alternative and complementary transcript profiling tool to GeneChip microarrays, especially in identifying novel genes and transcripts of low abundance.
Figures
Similar articles
-
Long versus short oligonucleotide microarrays for the study of gene expression in nonhuman primates.J Neurosci Methods. 2006 Apr 15;152(1-2):179-89. doi: 10.1016/j.jneumeth.2005.09.007. Epub 2005 Oct 25. J Neurosci Methods. 2006. PMID: 16253343
-
Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays.BMC Genomics. 2006 Mar 21;7:59. doi: 10.1186/1471-2164-7-59. BMC Genomics. 2006. PMID: 16551369 Free PMC article.
-
cDNA amplification by SMART-PCR and suppression subtractive hybridization (SSH)-PCR.Methods Mol Biol. 2009;496:223-43. doi: 10.1007/978-1-59745-553-4_15. Methods Mol Biol. 2009. PMID: 18839114 Review.
-
Development of a cDNA microarray of zebra mussel (Dreissena polymorpha) foot and its use in understanding the early stage of underwater adhesion.Gene. 2009 May 1;436(1-2):71-80. doi: 10.1016/j.gene.2009.01.007. Epub 2009 Jan 23. Gene. 2009. PMID: 19393183
-
[The methods of identifying differences in mRNA expression].Sheng Wu Gong Cheng Xue Bao. 2002 Jul;18(4):521-4. Sheng Wu Gong Cheng Xue Bao. 2002. PMID: 12385257 Review. Chinese.
Cited by
-
A genomic approach to investigate expression profiles in Slovenian patients with gastric cancer.Oncol Lett. 2011 Sep 1;2(5):1003-1014. doi: 10.3892/ol.2011.362. Epub 2011 Jul 7. Oncol Lett. 2011. PMID: 22866164 Free PMC article.
-
Identification of male gametogenesis expressed genes from the scallop Nodipecten subnodosus by suppressive subtraction hybridization and pyrosequencing.PLoS One. 2013 Sep 16;8(9):e73176. doi: 10.1371/journal.pone.0073176. eCollection 2013. PLoS One. 2013. PMID: 24066034 Free PMC article.
-
A Dehydration-Induced Eukaryotic Translation Initiation Factor iso4G Identified in a Slow Wilting Soybean Cultivar Enhances Abiotic Stress Tolerance in Arabidopsis.Front Plant Sci. 2018 Mar 2;9:262. doi: 10.3389/fpls.2018.00262. eCollection 2018. Front Plant Sci. 2018. PMID: 29552022 Free PMC article.
-
Spaceflight alters the gene expression profile of cervical cancer cells.Chin J Cancer. 2011 Dec;30(12):842-52. doi: 10.5732/cjc.011.10174. Epub 2011 Nov 18. Chin J Cancer. 2011. PMID: 22098948 Free PMC article.
-
Trichomonas vaginalis adherence mediates differential gene expression in human vaginal epithelial cells.Cell Microbiol. 2005 Jun;7(6):887-97. doi: 10.1111/j.1462-5822.2005.00522.x. Cell Microbiol. 2005. PMID: 15888089 Free PMC article.
References
-
- Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SP. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. - PubMed
-
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. - DOI - PMC - PubMed
-
- Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

