Computational protein biomarker prediction: a case study for prostate cancer
- PMID: 15113409
- PMCID: PMC406491
- DOI: 10.1186/1471-2105-5-26
Computational protein biomarker prediction: a case study for prostate cancer
Abstract
Background: Recent technological advances in mass spectrometry pose challenges in computational mathematics and statistics to process the mass spectral data into predictive models with clinical and biological significance. We discuss several classification-based approaches to finding protein biomarker candidates using protein profiles obtained via mass spectrometry, and we assess their statistical significance. Our overall goal is to implicate peaks that have a high likelihood of being biologically linked to a given disease state, and thus to narrow the search for biomarker candidates.
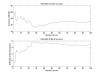
Results: Thorough cross-validation studies and randomization tests are performed on a prostate cancer dataset with over 300 patients, obtained at the Eastern Virginia Medical School using SELDI-TOF mass spectrometry. We obtain average classification accuracies of 87% on a four-group classification problem using a two-stage linear SVM-based procedure and just 13 peaks, with other methods performing comparably.
Conclusions: Modern feature selection and classification methods are powerful techniques for both the identification of biomarker candidates and the related problem of building predictive models from protein mass spectrometric profiles. Cross-validation and randomization are essential tools that must be performed carefully in order not to bias the results unfairly. However, only a biological validation and identification of the underlying proteins will ultimately confirm the actual value and power of any computational predictions.
Figures
References
-
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Series in Statistics Springer; 2001.
-
- Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. - PubMed
-
- Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

