Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes
- PMID: 15113441
- PMCID: PMC408462
- DOI: 10.1186/1471-2407-4-9
Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes
Abstract
Background: Subtle functional deficiencies in highly conserved DNA repair or growth regulatory processes resulting from polymorphic variation may increase genetic susceptibility to breast cancer. Polymorphisms in DNA repair genes can impact protein function leading to genomic instability facilitated by growth stimulation and increased cancer risk. Thus, 19 single nucleotide polymorphisms (SNPs) in eight genes involved in base excision repair (XRCC1, APEX, POLD1), BRCA1 protein interaction (BRIP1, ZNF350, BRCA2), and growth regulation (TGFss1, IGFBP3) were evaluated.
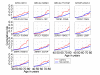
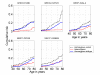
Methods: Genomic DNA samples were used in Taqman 5'-nuclease assays for most SNPs. Breast cancer risk to ages 50 and 70 were estimated using the kin-cohort method in which genotypes of relatives are inferred based on the known genotype of the index subject and Mendelian inheritance patterns. Family cancer history data was collected from a series of genotyped breast cancer cases (N = 748) identified within a cohort of female US radiologic technologists. Among 2,430 female first-degree relatives of cases, 190 breast cancers were reported.
Results: Genotypes associated with increased risk were: XRCC1 R194W (WW and RW vs. RR, cumulative risk up to age 70, risk ratio (RR) = 2.3; 95% CI 1.3-3.8); XRCC1 R399Q (QQ vs. RR, cumulative risk up to age 70, RR = 1.9; 1.1-3.9); and BRIP1 (or BACH1) P919S (SS vs. PP, cumulative risk up to age 50, RR = 6.9; 1.6-29.3). The risk for those heterozygous for BRCA2 N372H and APEX D148E were significantly lower than risks for homozygotes of either allele, and these were the only two results that remained significant after adjusting for multiple comparisons. No associations with breast cancer were observed for: APEX Q51H; XRCC1 R280H; IGFPB3 -202A>C; TGFss1 L10P, P25R, and T263I; BRCA2 N289H and T1915M; BRIP1 -64A>C; and ZNF350 (or ZBRK1) 1845C>T, L66P, R501S, and S472P.
Conclusion: Some variants in genes within the base-excision repair pathway (XRCC1) and BRCA1 interacting proteins (BRIP1) may play a role as low penetrance breast cancer risk alleles. Previous association studies of breast cancer and BRCA2 N372H and functional observations for APEX D148E ran counter to our findings of decreased risks. Due to the many comparisons, cautious interpretation and replication of these relationships are warranted.
Figures
Similar articles
-
Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from breast-ovarian cancer families and among early-onset breast cancer cases and reference individuals.Hum Mutat. 2003 Aug;22(2):121-8. doi: 10.1002/humu.10238. Hum Mutat. 2003. PMID: 12872252
-
Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer.Cancer Epidemiol Biomarkers Prev. 2006 Feb;15(2):353-8. doi: 10.1158/1055-9965.EPI-05-0653. Cancer Epidemiol Biomarkers Prev. 2006. PMID: 16492928
-
Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives.Breast Cancer Res Treat. 2009 May;115(1):51-5. doi: 10.1007/s10549-008-0052-z. Epub 2008 May 16. Breast Cancer Res Treat. 2009. PMID: 18483852
-
Breast cancer susceptibility: current knowledge and implications for genetic counselling.Eur J Hum Genet. 2009 Jun;17(6):722-31. doi: 10.1038/ejhg.2008.212. Epub 2008 Dec 17. Eur J Hum Genet. 2009. PMID: 19092773 Free PMC article. Review.
-
Symposium on 'The challenge of translating nutrition research into public health nutrition'. Session 2: Personalised nutrition. Genetic variation and disease risk: new advances.Proc Nutr Soc. 2009 May;68(2):113-21. doi: 10.1017/S0029665109001037. Epub 2009 Feb 11. Proc Nutr Soc. 2009. PMID: 19208270 Review.
Cited by
-
Age-dependent down-regulation of DNA polymerase δ1 in human lymphocytes.Mol Cell Biochem. 2012 Dec;371(1-2):157-63. doi: 10.1007/s11010-012-1432-6. Epub 2012 Aug 23. Mol Cell Biochem. 2012. PMID: 22915169
-
Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses.Hum Genet. 2006 May;119(4):376-88. doi: 10.1007/s00439-006-0135-z. Epub 2006 Feb 17. Hum Genet. 2006. PMID: 16485136
-
XRCC1 polymorphisms and breast cancer risk from the New York Site of the Breast Cancer Family Registry: A family-based case-control study.J Carcinog. 2010 Apr 16;9:4. doi: 10.4103/1477-3163.62535. J Carcinog. 2010. PMID: 20442803 Free PMC article.
-
Elevated expression of POLD1 is associated with poor prognosis in breast cancer.Oncol Lett. 2018 Nov;16(5):5591-5598. doi: 10.3892/ol.2018.9392. Epub 2018 Sep 4. Oncol Lett. 2018. PMID: 30344713 Free PMC article.
-
Insight into the roles of helicase motif Ia by characterizing Fanconi anemia group J protein (FANCJ) patient mutations.J Biol Chem. 2014 Apr 11;289(15):10551-10565. doi: 10.1074/jbc.M113.538892. Epub 2014 Feb 25. J Biol Chem. 2014. PMID: 24573678 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

