Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication
- PMID: 15113898
- PMCID: PMC400382
- DOI: 10.1128/jvi.78.10.5157-5169.2004
Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication
Abstract
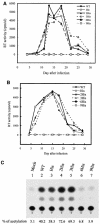
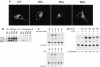
The biological significance of the presence of a long cytoplasmic domain in the envelope (Env) transmembrane protein gp41 of human immunodeficiency virus type 1 (HIV-1) is still not fully understood. Here we examined the effects of cytoplasmic tail elongation on virus replication and characterized the role of the C-terminal cytoplasmic tail in interactions with the Gag protein. Extensions with six and nine His residues but not with fewer than six His residues were found to severely inhibit virus replication through decreased Env electrophoretic mobility and reduced Env incorporation compared to the wild-type virus. These two mutants also exhibited distinct N glycosylation and reduced cell surface expression. An extension of six other residues had no deleterious effect on infectivity, even though some mutants showed reduced Env incorporation into the virus and/or decreased cell surface expression. We further show that these elongated cytoplasmic tails in a format of the glutathione S-transferase fusion protein still interacted effectively with the Gag protein. In addition, the immediate C terminus of the cytoplasmic tail was not directly involved in interactions with Gag, but the region containing the last 13 to 43 residues from the C terminus was critical for Env-Gag interactions. Taken together, our results demonstrate that HIV-1 Env can tolerate extension at its C terminus to a certain degree without loss of virus infectivity and Env-Gag interactions. However, extended elongation in the cytoplasmic tail may impair virus infectivity, Env cell surface expression, and Env incorporation into the virus.
Figures












References
-
- Celma, C. C., J. M. Manrique, J. L. Affranchino, E. Hunter, and S. A. Gonzalez. 2001. Domains in the simian immunodeficiency virus gp41 cytoplasmic tail required for envelope incorporation into particles. Virology 283:253-261. - PubMed
-
- Chen, S. S.-L., A. A. Ferrante, and E. F. Terwilliger. 1996. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology 226:260-268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

