Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants
- PMID: 15113899
- PMCID: PMC400349
- DOI: 10.1128/jvi.78.10.5170-5183.2004
Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants
Abstract
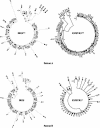
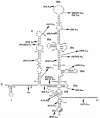
Reports of cerebral dysfunction in chronic hepatitis C virus (HCV) infection have led to the suggestion that HCV may infect the central nervous system (CNS). We used reverse transcription-PCR, cloning, and sequencing to define quasispecies for the HCV internal ribosomal entry site (IRES) and hypervariable region 1 (HVR1) in autopsy-derived brain, liver, lymph node, and serum samples. There was evidence of tissue compartmentalization of sequences in the brain in two patients, with between 24 and 55% of brain-derived IRES sequences absent from the serum, and significant phylogenetic and phenetic clustering of the brain and lymph node HVR1 sequences. The IRES initiates cap-independent translation of the viral polyprotein. Two unique brain-derived IRES mutations (C(204)-->A and G(243)-->A), which have previously been associated with lymphoid replication and altered translational efficiency in cell culture, were found in one patient. We used a dicistronic reporter vector to test whether brain-derived variants showed altered IRES-mediated translational efficiency, which might favor CNS infection. The translational efficiencies of the brain-derived IRES sequences were generally reduced compared to those of the master serum and liver sequences in rabbit reticulocyte cell lysates and two human cell lines, HuH7 (liver) and CHME3 (microglial). The C(204)-->A and G(243)-->A mutations showed preserved translational efficiency in HuH7 cells but reduced efficiency in CHME3 cells. Our data provide evidence that the CNS is a site of HCV replication, consistent with the recent demonstration of negative-strand HCV RNA in brain, and suggest that IRES polymorphisms may be important as a viral strategy of reduced translation to favor latency in the CNS.
Figures




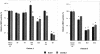
References
-
- Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, et al. 1992. The natural history of community-acquired hepatitis C in the United States. N. Engl. J. Med. 327:1899-1905. - PubMed
-
- Bouffard, P., P. H. Hayashi, R. Acevedo, N. Levy, and J. B. Zeldis. 1992. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J. Infect. Dis. 166:1276-1280. - PubMed
-
- Bronowicki, J. P., M. A. Loriot, V. Thiers, Y. Grignon, A. L. Zignego, and C. Brechot. 1998. Hepatitis C virus persistence in human hematopoietic cells injected into SCID mice. Hepatology 28:211-218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

