Analysis of host range phenotypes of primate hepadnaviruses by in vitro infections of hepatitis D virus pseudotypes
- PMID: 15113905
- PMCID: PMC400381
- DOI: 10.1128/jvi.78.10.5233-5243.2004
Analysis of host range phenotypes of primate hepadnaviruses by in vitro infections of hepatitis D virus pseudotypes
Abstract
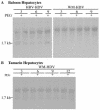
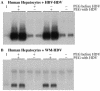
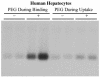
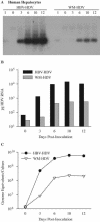
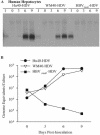
Hepatitis B virus (HBV) and woolly monkey hepatitis B virus (WMHBV) have natural host ranges that are limited to closely related species. The barrier for infection of primates seems to be at the adsorption and/or entry steps of the viral replication cycle, since a human hepatoma cell line is permissive for HBV and WMHBV replication following transfection of cloned DNA. We hypothesized that the HBV and WMHBV envelope proteins contain the principal viral determinants of host range. As previously shown by using the hepatitis D virus (HDV) system, recombinant HBV-HDV particles were infectious in chimpanzee as well as human hepatocytes. We extended the HDV system to include HDV particles pseudotyped with the WMHBV envelope. In agreement with the natural host ranges of HBV and WMHBV, in vitro infections demonstrated that HBV-HDV and WM-HDV particles preferentially infected human and spider monkey cells, respectively. Previous studies have implicated the pre-S1 region of the large (L) envelope protein in receptor binding and host range; therefore, recombinant HDV particles were pseudotyped with the hepadnaviral envelopes containing chimeric L proteins with the first 40 amino acids from the pre-S1 domain exchanged between HBV and WMHBV. Surprisingly, addition of the human amino terminus to the WMHBV L protein increased infectivity on spider monkey hepatocytes but did not increase infectivity for human hepatocytes. Based upon these data, we discuss the possibility that the L protein may be comprised of two domains that affect infectivity and that sequences downstream of residue 40 may influence host range and receptor binding or entry.
Figures




References
-
- Barrera, A., and R. E. Lanford. 2004. Infection of primary chimpanzee hepatocytes with recombinant hepatitis D virus particles: a surrogate model for hepatitis B virus. Methods Mol. Med. 95:131-142. - PubMed
-
- Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and size of the encapsidated HBV RNA. Virology 194:597-607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

