Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells
- PMID: 15117966
- PMCID: PMC2172041
- DOI: 10.1083/jcb.200310139
Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells
Abstract
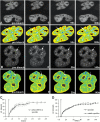
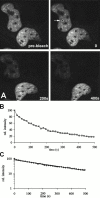
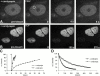
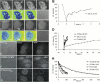
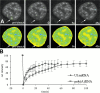
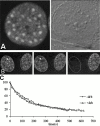
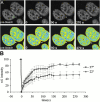
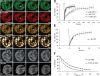
Many of the protein factors that play a role in nuclear export of mRNAs have been identified, but still little is known about how mRNAs are transported through the cell nucleus and which nuclear compartments are involved in mRNA transport. Using fluorescent 2'O-methyl oligoribonucleotide probes, we investigated the mobility of poly(A)+ RNA in the nucleoplasm and in nuclear speckles of U2OS cells. Quantitative analysis of diffusion using photobleaching techniques revealed that the majority of poly(A)+ RNA move throughout the nucleus, including in and out of speckles (also called SC-35 domains), which are enriched for splicing factors. Interestingly, in the presence of the transcription inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole, the association of poly(A)+ RNA with speckles remained dynamic. Our results show that RNA movement is energy dependent and that the proportion of nuclear poly(A)+ RNA that resides in speckles is a dynamic population that transiently interacts with speckles independent of the transcriptional status of the cell. Rather than the poly(A)+ RNA within speckles serving a stable structural role, our findings support the suggestion of a more active role of these regions in nuclear RNA metabolism and/or transport.
Figures
Similar articles
-
Tracking nuclear poly(A) RNA movement within and among speckle nuclear bodies and the surrounding nucleoplasm.Methods Mol Biol. 2013;1042:61-71. doi: 10.1007/978-1-62703-526-2_5. Methods Mol Biol. 2013. PMID: 23980000
-
In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains.J Cell Biol. 2006 Jan 30;172(3):373-81. doi: 10.1083/jcb.200503061. Epub 2006 Jan 23. J Cell Biol. 2006. PMID: 16431928 Free PMC article.
-
Nucleocytoplasmic transport of fluorescent mRNA in living mammalian cells: nuclear mRNA export is coupled to ongoing gene transcription.Genes Cells. 2006 Mar;11(3):305-17. doi: 10.1111/j.1365-2443.2006.00936.x. Genes Cells. 2006. PMID: 16483318
-
Nuclear speckles: a model for nuclear organelles.Nat Rev Mol Cell Biol. 2003 Aug;4(8):605-12. doi: 10.1038/nrm1172. Nat Rev Mol Cell Biol. 2003. PMID: 12923522 Review.
-
Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore.Curr Opin Cell Biol. 2005 Jun;17(3):294-301. doi: 10.1016/j.ceb.2005.04.007. Curr Opin Cell Biol. 2005. PMID: 15901500 Review.
Cited by
-
Onepot-Seq: capturing single-cell transcriptomes simultaneously in a continuous medium via transient localization of mRNA.Nucleic Acids Res. 2022 Dec 9;50(22):12621-12635. doi: 10.1093/nar/gkac665. Nucleic Acids Res. 2022. PMID: 35953080 Free PMC article.
-
Imaging intracellular RNA distribution and dynamics in living cells.Nat Methods. 2009 May;6(5):331-8. doi: 10.1038/nmeth.1321. Nat Methods. 2009. PMID: 19404252 Review.
-
Imaging gene expression in single living cells.Nat Rev Mol Cell Biol. 2004 Oct;5(10):855-61. doi: 10.1038/nrm1494. Nat Rev Mol Cell Biol. 2004. PMID: 15459666 Free PMC article. Review.
-
mRNA accumulation in the Cajal bodies of the diplotene larch microsporocyte.Chromosoma. 2012 Feb;121(1):37-48. doi: 10.1007/s00412-011-0339-4. Epub 2011 Sep 10. Chromosoma. 2012. PMID: 21909692 Free PMC article.
-
The dynamic pathway of nuclear RNA in eukaryotes.Nucleus. 2013 May-Jun;4(3):195-205. doi: 10.4161/nucl.24434. Epub 2013 Apr 11. Nucleus. 2013. PMID: 23580182 Free PMC article. Review.
References
-
- Bauren, G., and L. Wieslander. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 76:183–192. - PubMed
-
- Braun, I.C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536–20543. - PubMed
-
- Calado, C., and M. Carmo-Fonseca. 2000. Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J. Cell Sci. 113:2309–2318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

