Directed evolution of protein enzymes using nonhomologous random recombination
- PMID: 15118093
- PMCID: PMC406457
- DOI: 10.1073/pnas.0402202101
Directed evolution of protein enzymes using nonhomologous random recombination
Abstract
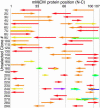
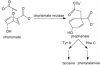
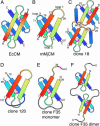
We recently reported the development of nonhomologous random recombination (NRR) as a method for nucleic acid diversification and applied NRR to the evolution of DNA aptamers. Here, we describe a modified method, protein NRR, that enables proteins to access diversity previously difficult or impossible to generate. We investigated the structural plasticity of protein folds and the ability of helical motifs to function in different contexts by applying protein NRR and in vivo selection to the evolution of chorismate mutase (CM) enzymes. Functional CM mutants evolved using protein NRR contained many insertions, deletions, and rearrangements. The distribution of these changes was not random but clustered in certain regions of the protein. Topologically rearranged but functional enzymes also emerged from these studies, indicating that multiple connectivities can accommodate a functional CM active site and demonstrating the ability to generate new domain connectivities through protein NRR. Protein NRR was also used to randomly recombine CM and fumarase, an unrelated but also alpha-helical protein. Whereas the resulting library contained fumarase fragments in many contexts before functional selection, library members surviving selection for CM activity invariably contained a CM core with fumarase sequences found only at the termini or in one loop. These results imply that internal helical fragments cannot be swapped between these proteins without the loss of nearly all CM activity. Our findings suggest that protein NRR will be useful in probing the functional requirements of enzymes and in the creation of new protein topologies.
Figures



Similar articles
-
Nucleic acid evolution and minimization by nonhomologous random recombination.Nat Biotechnol. 2002 Oct;20(10):1024-9. doi: 10.1038/nbt736. Epub 2002 Sep 9. Nat Biotechnol. 2002. PMID: 12219078 Free PMC article.
-
Design, selection, and characterization of a split chorismate mutase.Protein Sci. 2010 May;19(5):1000-10. doi: 10.1002/pro.377. Protein Sci. 2010. PMID: 20306491 Free PMC article.
-
Relative tolerance of mesostable and thermostable protein homologs to extensive mutation.Proteins. 2007 Feb 1;66(2):500-6. doi: 10.1002/prot.21227. Proteins. 2007. PMID: 17096428
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
-
Genetic selection as a tool in mechanistic enzymology and protein design.Ernst Schering Res Found Workshop. 2000;(32):253-68. doi: 10.1007/978-3-662-04042-3_9. Ernst Schering Res Found Workshop. 2000. PMID: 11077612 Review. No abstract available.
Cited by
-
Domain-swapping of mesophilic xylanase with hyper-thermophilic glucanase.BMC Biotechnol. 2012 Jun 7;12:28. doi: 10.1186/1472-6750-12-28. BMC Biotechnol. 2012. PMID: 22676349 Free PMC article.
-
Identification of eukaryotic promoter regulatory elements using nonhomologous random recombination.Nucleic Acids Res. 2007;35(17):5851-60. doi: 10.1093/nar/gkm634. Epub 2007 Aug 24. Nucleic Acids Res. 2007. PMID: 17720707 Free PMC article.
-
Discovery of a mRNA mitochondrial localization element in Saccharomyces cerevisiae by nonhomologous random recombination and in vivo selection.Nucleic Acids Res. 2007;35(20):6750-61. doi: 10.1093/nar/gkm777. Epub 2007 Oct 4. Nucleic Acids Res. 2007. PMID: 17916575 Free PMC article.
-
Recombination of protein fragments: a promising approach toward engineering proteins with novel nanomechanical properties.Protein Sci. 2008 Oct;17(10):1815-26. doi: 10.1110/ps.036376.108. Epub 2008 Jul 14. Protein Sci. 2008. PMID: 18628239 Free PMC article.
-
Simultaneous optimization of enzyme activity and quaternary structure by directed evolution.Protein Sci. 2005 Aug;14(8):2103-14. doi: 10.1110/ps.051431605. Epub 2005 Jun 29. Protein Sci. 2005. PMID: 15987889 Free PMC article.
References
-
- Wang, L., Brock, A., Herberich, B. & Schultz, P. G. (2001) Science 292, 498-500. - PubMed
-
- Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. (1996) Nat. Biotechnol. 14, 315-319. - PubMed
-
- Stemmer, W. P. (1994) Nature 370, 389-391. - PubMed
-
- Lim, W. A. & Sauer, R. T. (1989) Nature 339, 31-36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

