Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry
- PMID: 15118107
- PMCID: PMC406438
- DOI: 10.1073/pnas.0400873101
Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry
Abstract
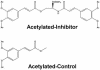
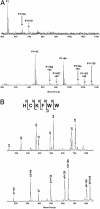
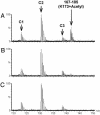
We report a methodology that combines affinity acetylation with MS analysis for accurate mapping of an inhibitor-binding site to a target protein. For this purpose, we used a known HIV-1 integrase inhibitor containing aryl di-O-acetyl groups (Acetylated-Inhibitor). In addition, we designed a control compound (Acetylated-Control) that also contained an aryl di-O-acetyl group but did not inhibit HIV-1 integrase. Examination of the reactivity of these compounds with a model peptide library, which collectively contained all 20 natural amino acids, revealed that aryl di-O-acetyl compounds effectively acetylate Cys, Lys, and Tyr residues. Acetylated-Inhibitor and Acetylated-Control exhibited comparable chemical reactivity with respect to these small peptides. However, these two compounds differed markedly in their interactions with HIV-1 integrase. In particular, Acetylated-Inhibitor specifically acetylated K173 at its inhibitory concentration (3 microM) whereas this site remained unrecognized by Acetylated-Control. Our data enabled creation of a detailed model for the integrase:Acetylated-Inhibitor complex, which indicated that the inhibitor selectively binds at an architecturally critical region of the protein. The methodology reported herein has a generic application for systems involving a variety of ligand-protein interactions.
Figures



Similar articles
-
Triketoacid inhibitors of HIV-integrase: a new chemotype useful for probing the integrase pharmacophore.Bioorg Med Chem Lett. 2006 Jun 1;16(11):2920-4. doi: 10.1016/j.bmcl.2006.03.010. Epub 2006 Mar 20. Bioorg Med Chem Lett. 2006. PMID: 16546383
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
Hyrtiosal, from the marine sponge Hyrtios erectus, inhibits HIV-1 integrase binding to viral DNA by a new inhibitor binding site.ChemMedChem. 2008 Jan;3(1):173-80. doi: 10.1002/cmdc.200700223. ChemMedChem. 2008. PMID: 17943714
-
Study on the inhibitory mechanism and binding mode of the hydroxycoumarin compound NSC158393 to HIV-1 integrase by molecular modeling.Biopolymers. 2009 Sep;91(9):700-9. doi: 10.1002/bip.21211. Biopolymers. 2009. PMID: 19382173
-
New approaches for inhibiting HIV integrase: a journey beyond the active site.Curr Opin Investig Drugs. 2009 Feb;10(2):129-36. Curr Opin Investig Drugs. 2009. PMID: 19197790 Review.
Cited by
-
Discovery of a small-molecule HIV-1 integrase inhibitor-binding site.Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):10080-5. doi: 10.1073/pnas.0511254103. Epub 2006 Jun 19. Proc Natl Acad Sci U S A. 2006. PMID: 16785440 Free PMC article.
-
Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors.Expert Rev Mol Med. 2013 Nov 26;15:e14. doi: 10.1017/erm.2013.15. Expert Rev Mol Med. 2013. PMID: 24274067 Free PMC article. Review.
-
Allosteric inhibitor development targeting HIV-1 integrase.ChemMedChem. 2011 Feb 7;6(2):228-41. doi: 10.1002/cmdc.201000443. Epub 2011 Jan 12. ChemMedChem. 2011. PMID: 21275045 Free PMC article. Review.
-
5-Nitro-3-(2-(4-phenylthiazol-2-yl)hydrazineylidene)indolin-2-one derivatives inhibit HIV-1 replication by a multitarget mechanism of action.Front Cell Infect Microbiol. 2023 Jun 23;13:1193280. doi: 10.3389/fcimb.2023.1193280. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37424782 Free PMC article.
-
Methods for the Analyses of Inhibitor-Induced Aberrant Multimerization of HIV-1 Integrase.Methods Mol Biol. 2016;1354:149-64. doi: 10.1007/978-1-4939-3046-3_10. Methods Mol Biol. 2016. PMID: 26714710 Free PMC article.
References
-
- Brown, P. O. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 161-204.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

