Polypyrimidine tract binding protein modulates efficiency of polyadenylation
- PMID: 15121839
- PMCID: PMC400487
- DOI: 10.1128/MCB.24.10.4174-4183.2004
Polypyrimidine tract binding protein modulates efficiency of polyadenylation
Erratum in
- Mol Cell Biol. 2004 Aug;24(15):6889
Abstract
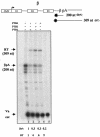
Polypyrimidine tract binding protein (PTB) is a major hnRNP protein with multiple roles in mRNA metabolism, including regulation of alternative splicing and internal ribosome entry site-driven translation. We show here that a fourfold overexpression of PTB results in a 75% reduction of mRNA levels produced from transfected gene constructs with different polyadenylation signals (pA signals). This effect is due to the reduced efficiency of mRNA 3' end cleavage, and in vitro analysis reveals that PTB competes with CstF for recognition of the pA signal's pyrimidine-rich downstream sequence element. This may be analogous to its role in alternative splicing, where PTB competes with U2AF for binding to pyrimidine-rich intronic sequences. The pA signal of the C2 complement gene unusually possesses a PTB-dependent upstream sequence, so that knockdown of PTB expression by RNA interference reduces C2 mRNA expression even though PTB overexpression still inhibits polyadenylation. Consequently, we show that PTB can act as a regulator of mRNA expression through both its negative and positive effects on mRNA 3' end processing.
Figures



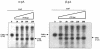


References
-
- Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. - PubMed
-
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
