The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres
- PMID: 15121850
- PMCID: PMC400474
- DOI: 10.1128/MCB.24.10.4309-4320.2004
The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres
Abstract
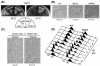
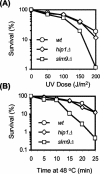
HIRA-like (Hir) proteins are evolutionarily conserved and are implicated in the assembly of repressive chromatin. In Saccharomyces cerevisiae, Hir proteins contribute to the function of centromeres. However, S. cerevisiae has point centromeres that are structurally different from the complex centromeres of metazoans. In contrast, Schizosaccharomyces pombe has complex centromeres whose domain structure is conserved with that of human centromeres. Therefore, we examined the functions of the fission yeast Hir proteins Slm9 and the previously uncharacterised protein Hip1. Deletion of hip1(+) resulted in phenotypes that were similar to those described previously for slm9 Delta cells: a cell cycle delay, synthetic lethality with cdc25-22, and poor recovery from nitrogen starvation. However, while it has previously been shown that Slm9 is not required for the periodic expression of histone H2A, we found that loss of Hip1 led to derepression of core histone genes expression outside of S phase. Importantly, we found that deletion of either hip1(+) or slm9(+) resulted in increased rates of chromosome loss, increased sensitivity to spindle damage, and reduced transcriptional silencing in the outer centromeric repeats. Thus, S. pombe Hir proteins contribute to pericentromeric heterochromatin, and our data thus suggest that Hir proteins may be required for the function of metazoan centromeres.
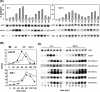
Figures

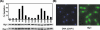

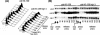



References
-
- Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. - PubMed
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, 3rd, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
-
- Basi, G., E. Schmid, and K. Maundrell. 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
