Hip1-related mutant mice grow and develop normally but have accelerated spinal abnormalities and dwarfism in the absence of HIP1
- PMID: 15121852
- PMCID: PMC400480
- DOI: 10.1128/MCB.24.10.4329-4340.2004
Hip1-related mutant mice grow and develop normally but have accelerated spinal abnormalities and dwarfism in the absence of HIP1
Abstract
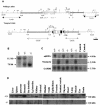
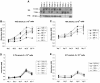
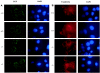
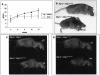
In mice and humans, there are two known members of the Huntingtin interacting protein 1 (HIP1) family, HIP1 and HIP1-related (HIP1r). Based on structural and functional data, these proteins participate in the clathrin trafficking network. The inactivation of Hip1 in mice leads to spinal, hematopoietic, and testicular defects. To investigate the biological function of HIP1r, we generated a Hip1r mutant allele in mice. Hip1r homozygous mutant mice are viable and fertile without obvious morphological abnormalities. In addition, embryonic fibroblasts derived from these mice do not have gross abnormalities in survival, proliferation, or clathrin trafficking pathways. Altogether, this demonstrates that HIP1r is not necessary for normal development of the embryo or for normal adulthood and suggests that HIP1 or other functionally related members of the clathrin trafficking network can compensate for HIP1r absence. To test the latter, we generated mice deficient in both HIP1 and HIP1r. These mice have accelerated development of abnormalities seen in Hip1 -deficient mice, including kypholordosis and growth defects. The severity of the Hip1r/Hip1 double-knockout phenotype compared to the Hip1 knockout indicates that HIP1r partially compensates for HIP1 function in the absence of HIP1 expression, providing strong evidence that HIP1 and HIP1r have overlapping roles in vivo.
Figures

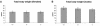


References
-
- Anonymous. 2004. Mouse genome database. Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine. [Online.] http://www.informatics.jax.org.
-
- Chopra, V. S., M. Metzler, D. M. Rasper, A. E. Engqvist-Goldstein, R. Singaraja, L. Gan, K. M. Fichter, K. McCutcheon, D. Drubin, D. W. Nicholson, and M. R. Hayden. 2000. HIP12 is a non-proapoptotic member of a gene family including HIP1, an interacting protein with Huntingtin. Mamm. Genome 11: 1006-1015. - PubMed
-
- Dlugosz, A. A., A. B. Glick, T. Tennenbaum, W. C. Weinberg, and S. H. Yuspa. 1995. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 254: 3-20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
