TRAF family proteins link PKR with NF-kappa B activation
- PMID: 15121867
- PMCID: PMC400457
- DOI: 10.1128/MCB.24.10.4502-4512.2004
TRAF family proteins link PKR with NF-kappa B activation
Abstract
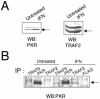
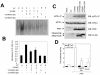
The double-stranded RNA (dsRNA)-dependent protein kinase PKR activates NF-kappa B via the I kappa B kinase (IKK) complex, but little is known about additional molecules that may be involved in this pathway. Analysis of the PKR sequence enabled us to identify two putative TRAF-interacting motifs. The viability of such an interaction was further suggested by computer modeling. Here, we present evidence of the colocalization and physical interaction between PKR and TRAF family proteins in vivo, as shown by immunoprecipitation and confocal microscopy experiments. This interaction is induced upon PKR dimerization. Most importantly, we show that the binding between PKR and TRAFs is functionally relevant, as observed by the absence of NF-kappa B activity upon PKR expression in cells genetically deficient in TRAF2 and TRAF5 or after expression of TRAF dominant negative molecules. On the basis of sequence information and mutational and computer docking analyses, we favored a TRAF-PKR interaction model in which the C-terminal domain of TRAF binds to a predicted TRAF interaction motif present in the PKR kinase domain. Altogether, our data suggest that TRAF family proteins are key components located downstream of PKR that have an important role in mediating activation of NF-kappa B by the dsRNA-dependent protein kinase.
Figures







References
-
- Akiba, H., H. Nakano, S. Nishinaka, M. Shindo, T. Kobata, M. Atsuta, C. Morimoto, C. F. Ware, N. L. Malinin, D. Wallach, H. Yagita, and K. Okumura. 1998. CD27, a member of the tumor necrosis factor receptor superfamily, activates NF-κB and stress-activated protein kinase/c-Jun N-terminal kinase via TRAF2, TRAF5 and NF-κB-inducing kinase. J. Biol. Chem. 273:13353-13358. - PubMed
-
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. - PubMed
-
- Arch, R. H., R. W. Gedrich, and C. B. Thompson. 1998. Tumor necrosis factor receptor-associated factors (TRAFs)—a family of adapter proteins that regulates life and death. Genes Dev. 12:2821-2830. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
