DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene
- PMID: 15121897
- PMCID: PMC419437
- DOI: 10.1093/nar/gkh527
DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene
Abstract
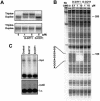
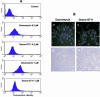
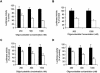
Triplex-forming oligonucleotides (TFO) that bind DNA in a sequence-specific manner might be used as selective repressors of gene expression and gene-targeted therapeutics. However, many factors, including instability of triple helical complexes in cells, limit the efficacy of this approach. In the present study, we tested whether covalent linkage of a TFO to daunomycin, which is a potent DNA-intercalating agent and anticancer drug, could increase stability of the triple helix and activity of the oligonucleotide in cells. The 11mer daunomycin-conjugated GT (dauno-GT11) TFO targeted a sequence upstream of the P2 promoter, a site known to be critical for transcription of the c-myc gene. Band-shift assays showed that the dauno-GT11 formed triplex DNA with enhanced stability compared to the unmodified TFO. Band shift and footprinting experiments demonstrated that binding of dauno-GT11 was highly sequence-specific with exclusive binding to the 11 bp target site in the c-myc promoter. The daunomycin-conjugated TFO inhibited transcription in vitro and reduced c-myc promoter activity in prostate and breast cancer cells. The daunomycin-conjugated TFO was taken up by cells with a distinctive intracellular distribution compared to free daunomycin. However, cationic lipid-mediated delivery was required for enhanced cellular uptake, nuclear localization and biological activity of the TFO in cells. Dauno-GT11 reduced transcription of the endogenous c-myc gene in cells, but did not affect expression of non-target genes, such as ets-1 and ets-2, which contained very similar target sequences in their promoters. Daunomycin-conjugated control oligonucleotides unable to form triplex DNA with the target sequence did not have any effect in these assays, indicating that daunomycin was not directly responsible for the activity of daunomycin-conjugated TFO. Thus, attachment of daunomycin resulted in increased triplex stability and biological activity of the 11mer GT-rich TFO without compromising its specificity. These results encourage further testing of this approach to develop novel antigene therapeutics.
Figures




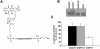

References
-
- Chan P.P. and Glazer,P.M. (1997) Triplex DNA: fundamentals, advances and potential applications for gene therapy. Mol. Med., 75, 267–282. - PubMed
-
- Praseuth D., Guieysse,A.L. and Helene,C. (1999) Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta, 1489, 181–206. - PubMed
-
- Casey B.P. and Glazer,P.M. (2001) Gene targeting via triple-helix formation. Prog. Nucleic Acid Res. Mol. Biol., 67, 163–192. - PubMed
-
- Winters T.A. (2000) Gene targeted agents: new opportunities for rational drug development. Curr. Opin. Mol. Ther., 2, 670–681. - PubMed
-
- Moser H.E. and Dervan,P.B. (1987) Sequence-specific cleavage of double helical DNA by triple helix formation. Science, 238, 645–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

