A versatile assay for high-throughput gene expression profiling on universal array matrices
- PMID: 15123585
- PMCID: PMC479115
- DOI: 10.1101/gr.2167504
A versatile assay for high-throughput gene expression profiling on universal array matrices
Abstract
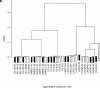
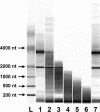
We report a flexible, sensitive, and quantitative gene-expression profiling system for assaying more than 400 genes, with three probes per gene, for 96 samples in parallel. The cDNA-mediated annealing, selection, extension and ligation (DASL) assay targets specific transcripts, using oligonucleotides containing unique address sequences that can hybridize to universal arrays. Cell-specific gene expression profiles were obtained using this assay for hormone-treated cell lines and laser-capture microdissected cancer tissues. Gene expression profiles derived from this assay were consistent with those determined by qRT-PCR. The DASL assay has been automated for use with a bead-based 96-array matrix system. The combined high-throughput assay and readout system is accurate and efficient, and can cost-effectively profile the expression of hundreds of genes in thousands of samples.
Figures





References
-
- Barker, D.L., Theriault, G., Che, D., Dickinson, T. Shen, R., and Kain, R. 2003. Self-assembled random arrays: High-performance imaging and genomics applications on a high-density microarray platform. Proc. SPIE 4966: 1-11.
-
- Chuaqui, R.F., Bonner, R.F., Best, C.J., Gillespie, J.W., Flaig, M.J., Hewitt, S.M., Phillips, J.L., Krizman, D.B., Tangrea, M.A., Ahram, M., et al. 2002. Post-analysis follow-up and validation of microarray experiments. Nat. Genet. 32: S509-S514. - PubMed
-
- Dhanasekaran, S.M., Barrette, T.R., Ghosh, D., Shah, R., Varambally, S., Kurachi, K., Pienta, K.J., Rubin, M.A., and Chinnaiyan, A.M. 2001. Delineation of prognostic biomarkers in prostate cancer. Nature 412: 822-826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
