Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel
- PMID: 15123587
- PMCID: PMC479118
- DOI: 10.1101/gr.1949704
Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel
Abstract
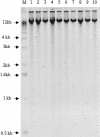
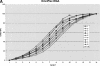
Comprehensive genome scans involving many thousands of SNP assays will require significant amounts of genomic DNA from each sample. We report two successful methods for amplifying whole-genomic DNA prior to SNP analysis, multiple displacement amplification, and OmniPlex technology. We determined the coverage of amplification by analyzing a SNP linkage marker set that contained 2320 SNP markers spread across the genome at an average distance of 2.5 cM. We observed a concordance of >99.8% in genotyping results from genomic DNA and amplified DNA, strongly indicating the ability of both methods used to amplify genomic DNA in a highly representative manner. Furthermore, we were able to achieve a SNP call rate of >98% in both genomic and amplified DNA. The combination of whole-genome amplification and comprehensive SNP linkage analysis offers new opportunities for genetic analysis in clinical trials, disease association studies, and archiving of DNA samples.
Figures


References
-
- Barker, D.L., Theriault, G., Che, D., Dickinson, T., Shen, R., and Kain, R. 2003. Self-assembled random arrays: High-performance imaging and genomics applications on a high-density microarray platform. Proc. SPIE 4966: 1-11.
-
- Buckley, P.G., Mantripragada, K.K., Benetkiewicz, M., Tapia-Paez, I., Diaz De Stahl, T., Rosenquist, M., Ali, H., Jarbo, C., De Bustos, C., Hirvela, C., et al. 2002. A full-coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum. Mol. Genet. 11: 3221-3229. - PubMed
WEB SITE REFERENCES
-
- www.rubicongenomics.com; Contains information on Omniplex DNA Amplification Kits.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
