The Ensembl analysis pipeline
- PMID: 15123589
- PMCID: PMC479123
- DOI: 10.1101/gr.1859804
The Ensembl analysis pipeline
Abstract
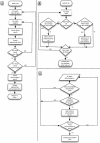
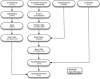
The Ensembl pipeline is an extension to the Ensembl system which allows automated annotation of genomic sequence. The software comprises two parts. First, there is a set of Perl modules ("Runnables" and "RunnableDBs") which are 'wrappers' for a variety of commonly used analysis tools. These retrieve sequence data from a relational database, run the analysis, and write the results back to the database. They inherit from a common interface, which simplifies the writing of new wrapper modules. On top of this sits a job submission system (the "RuleManager") which allows efficient and reliable submission of large numbers of jobs to a compute farm. Here we describe the fundamental software components of the pipeline, and we also highlight some features of the Sanger installation which were necessary to enable the pipeline to scale to whole-genome analysis.
Figures

References
-
- Durbin, R. and Mieg, T. 1991. A C. elegans Database. Documentation, code and data available from anonymous FTP servers at http://lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk and ncbi.nlm.nih.gov.
WEB SITE REFERENCES
-
- http://www.acedb.org; AceDB.
-
- http://www.platform.com/products/LSF; Load Sharing Facility.
-
- http://www.ncbi.nlm.nih.gov/genome/guide/build.html#annot; NCBI's annotation pipeline.
-
- http://vega.sanger.ac.uk; the Vertebrate Genome Annotation database.
-
- http://cvsweb.sanger.ac.uk; Public CVS repository for the Ensembl software.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
