The Ensembl automatic gene annotation system
- PMID: 15123590
- PMCID: PMC479124
- DOI: 10.1101/gr.1858004
The Ensembl automatic gene annotation system
Abstract
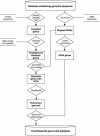
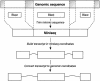
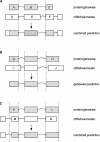
As more genomes are sequenced, there is an increasing need for automated first-pass annotation which allows timely access to important genomic information. The Ensembl gene-building system enables fast automated annotation of eukaryotic genomes. It annotates genes based on evidence derived from known protein, cDNA, and EST sequences. The gene-building system rests on top of the core Ensembl (MySQL) database schema and Perl Application Programming Interface (API), and the data generated are accessible through the Ensembl genome browser (http://www.ensembl.org). To date, the Ensembl predicted gene sets are available for the A. gambiae, C. briggsae, zebrafish, mouse, rat, and human genomes and have been heavily relied upon in the publication of the human, mouse, rat, and A. gambiae genome sequence analysis. Here we describe in detail the gene-building system and the algorithms involved. All code and data are freely available from http://www.ensembl.org.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
