Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core
- PMID: 15123674
- PMCID: PMC7982547
- DOI: 10.1074/jbc.M403760200
Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core
Abstract
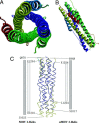
The surface transmembrane glycoprotein is responsible for mediating virion attachment to cell and subsequent virus-cell membrane fusion. However, the molecular mechanisms for the viral entry of coronaviruses remain poorly understood. The crystal structure of the fusion core of mouse hepatitis virus S protein, which represents the first fusion core structure of any coronavirus, reveals a central hydrophobic coiled coil trimer surrounded by three helices in an oblique, antiparallel manner. This structure shares significant similarity with both the low pH-induced conformation of influenza hemagglutinin and fusion core of HIV gp41, indicating that the structure represents a fusion-active state formed after several conformational changes. Our results also indicate that the mechanisms for the viral fusion of coronaviruses are similar to those of influenza virus and HIV. The coiled coil structure has unique features, which are different from other viral fusion cores. Highly conserved heptad repeat 1 (HR1) and HR2 regions in coronavirus spike proteins indicate a similar three-dimensional structure among these fusion cores and common mechanisms for the viral fusion. We have proposed the binding regions of HR1 and HR2 of other coronaviruses and a structure model of their fusion core based on our mouse hepatitis virus fusion core structure and sequence alignment. Drug discovery strategies aimed at inhibiting viral entry by blocking hairpin formation may be applied to the inhibition of a number of emerging infectious diseases, including severe acute respiratory syndrome.
Figures



Similar articles
-
Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core.J Biol Chem. 2004 Nov 19;279(47):49414-9. doi: 10.1074/jbc.M408782200. Epub 2004 Sep 1. J Biol Chem. 2004. PMID: 15345712 Free PMC article.
-
Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein.Proc Natl Acad Sci U S A. 2004 Dec 28;101(52):17958-63. doi: 10.1073/pnas.0406128102. Epub 2004 Dec 16. Proc Natl Acad Sci U S A. 2004. PMID: 15604146 Free PMC article.
-
Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555
-
Mechanisms of coronavirus cell entry mediated by the viral spike protein.Viruses. 2012 Jun;4(6):1011-33. doi: 10.3390/v4061011. Epub 2012 Jun 20. Viruses. 2012. PMID: 22816037 Free PMC article. Review.
-
Receptor specificity and receptor-induced conformational changes in mouse hepatitis virus spike glycoprotein.Adv Exp Med Biol. 2001;494:173-81. doi: 10.1007/978-1-4615-1325-4_29. Adv Exp Med Biol. 2001. PMID: 11774465 Review. No abstract available.
Cited by
-
Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry.J Virol. 2006 Feb;80(3):1302-10. doi: 10.1128/JVI.80.3.1302-1310.2006. J Virol. 2006. PMID: 16415007 Free PMC article.
-
Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine.Biochem Biophys Res Commun. 2004 Nov 12;324(2):773-81. doi: 10.1016/j.bbrc.2004.09.106. Biochem Biophys Res Commun. 2004. PMID: 15474494 Free PMC article.
-
Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion.J Virol. 2008 Feb;82(3):1414-24. doi: 10.1128/JVI.01674-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032498 Free PMC article.
-
Exploitation of viral properties for intracellular delivery.J Pept Sci. 2014 Jul;20(7):468-78. doi: 10.1002/psc.2649. Epub 2014 May 30. J Pept Sci. 2014. PMID: 24889153 Free PMC article. Review.
-
gH625: a milestone in understanding the many roles of membranotropic peptides.Biochim Biophys Acta. 2015 Jan;1848(1 Pt A):16-25. doi: 10.1016/j.bbamem.2014.10.006. Epub 2014 Oct 12. Biochim Biophys Acta. 2015. PMID: 25305339 Free PMC article. Review.
References
-
- Spaan W., Cavanagh D., Horzinek M.C. J. Gen. Virol. 1988;69:2939–2952. - PubMed
-
- Siddell S., Wege H., Ter Meulen V. J. Gen. Virol. 1983;64:761–776. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

