The cell surface receptor SLAM controls T cell and macrophage functions
- PMID: 15123745
- PMCID: PMC2211908
- DOI: 10.1084/jem.20031835
The cell surface receptor SLAM controls T cell and macrophage functions
Abstract
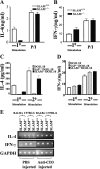
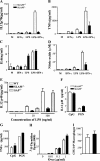
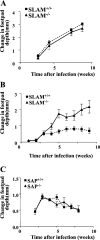
Signaling lymphocyte activation molecule (SLAM), a glycoprotein expressed on activated lymphocytes and antigen-presenting cells, has been shown to be a coregulator of antigen-driven T cell responses and is one of the two receptors for measles virus. Here we show that T cell receptor-induced interleukin (IL)-4 secretion by SLAM(-/-) CD4(+) cells is down-regulated, whereas interferon gamma production by CD4(+) T cells is only slightly up-regulated. Although SLAM controls production of IL-12, tumor necrosis factor, and nitric oxide in response to lipopolysaccharide (LPS) by macrophages, SLAM does not regulate phagocytosis and responses to peptidoglycan or CpG. Thus, SLAM acts as a coreceptor that regulates signals transduced by the major LPS receptor Toll-like receptor 4 on the surface of mouse macrophages. A defective macrophage function resulted in an inability of SLAM(-/-) C57Bl/6 mice to remove the parasite Leishmania major. We conclude that the coreceptor SLAM plays a central role at the interface of acquired and innate immune responses.
Figures

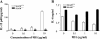
References
-
- Cocks, B.G., C.C. Chang, J.M. Carballido, H. Yssel, J.E. de Vries, and G. Aversa. 1995. A novel receptor involved in T-cell activation. Nature. 376:260–263. - PubMed
-
- Engel, P., M.J. Eck, and C. Terhorst. 2003. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat. Rev. Immunol. 3:813–821. - PubMed
-
- Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 406:893–897. - PubMed
-
- Sayos, J., C. Wu, M. Morra, N. Wang, X. Zhang, D. Allen, S. van Schaik, L. Notarangelo, R. Geha, M.G. Roncarolo, et al. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 395:462–469. - PubMed
-
- Latour, S., G. Gish, C.D. Helgason, R.K. Humphries, T. Pawson, and A. Veillette. 2001. Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product. Nat. Immunol. 2:681–690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

