The diploid genome sequence of Candida albicans
- PMID: 15123810
- PMCID: PMC409918
- DOI: 10.1073/pnas.0401648101
The diploid genome sequence of Candida albicans
Abstract
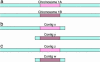
We present the diploid genome sequence of the fungal pathogen Candida albicans. Because C. albicans has no known haploid or homozygous form, sequencing was performed as a whole-genome shotgun of the heterozygous diploid genome in strain SC5314, a clinical isolate that is the parent of strains widely used for molecular analysis. We developed computational methods to assemble a diploid genome sequence in good agreement with available physical mapping data. We provide a whole-genome description of heterozygosity in the organism. Comparative genomic analyses provide important clues about the evolution of the species and its mechanisms of pathogenesis.
Figures


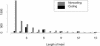

References
-
- Poulter, R. T. (1987) Crit. Rev. Microbiol. 15, 97-101. - PubMed
-
- Chibana, H., Beckerman, J. L. & Magee, P. T. (2000) Genome Res. 10, 1865-1877. - PubMed
-
- Hull, C. M., Raisner, R. M. & Johnson, A. D. (2000) Science 289, 307-310. - PubMed
-
- Magee, B. B. & Magee, P. T. (2000) Science 289, 310-313. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

