Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure
- PMID: 15123812
- PMCID: PMC409911
- DOI: 10.1073/pnas.0401799101
Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure
Abstract
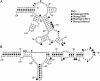
A dynamic programming algorithm for prediction of RNA secondary structure has been revised to accommodate folding constraints determined by chemical modification and to include free energy increments for coaxial stacking of helices when they are either adjacent or separated by a single mismatch. Furthermore, free energy parameters are revised to account for recent experimental results for terminal mismatches and hairpin, bulge, internal, and multibranch loops. To demonstrate the applicability of this method, in vivo modification was performed on 5S rRNA in both Escherichia coli and Candida albicans with 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate, dimethyl sulfate, and kethoxal. The percentage of known base pairs in the predicted structure increased from 26.3% to 86.8% for the E. coli sequence by using modification constraints. For C. albicans, the accuracy remained 87.5% both with and without modification data. On average, for these sequences and a set of 14 sequences with known secondary structure and chemical modification data taken from the literature, accuracy improves from 67% to 76%. This enhancement primarily reflects improvement for three sequences that are predicted with <40% accuracy on the basis of energetics alone. For these sequences, inclusion of chemical modification constraints improves the average accuracy from 28% to 78%. For the 11 sequences with <6% pseudoknotted base pairs, structures predicted with constraints from chemical modification contain on average 84% of known canonical base pairs.
Figures
References
-
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. - PubMed
-
- Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. - PubMed
-
- Cullen, B. R. (2002) Nat. Immunol. 3, 597-599. - PubMed
-
- McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737-747. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

