OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene
- PMID: 15126450
- PMCID: PMC400630
- DOI: 10.1128/JB.186.10.2909-2920.2004
OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene
Abstract
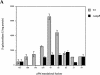
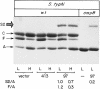
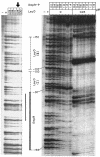
The Salmonella enterica serovar Typhi ompS2 gene codes for a 362-amino-acid outer membrane protein that contains motifs common to the porin superfamily. It is expressed at very low levels compared to the major OmpC and OmpF porins, as observed for S. enterica serovar Typhi OmpS1, Escherichia coli OmpN, and Klebsiella pneumoniae OmpK37 quiescent porins. A region of 316 bp, between nucleotides -413 and -97 upstream of the transcriptional start point, is involved in negative regulation, as its removal resulted in a 10-fold increase in ompS2 expression in an S. enterica serovar Typhi wild-type strain. This enhancement in expression was not observed in isogenic mutant strains, which had specific deletions of the regulatory ompB (ompR envZ) operon. Furthermore, ompS2 expression was substantially reduced in the presence of the OmpR D55A mutant, altered in the major phosphorylation site. Upon random mutagenesis, a mutant where the transposon had inserted into the upstream regulatory region of the gene coding for the LeuO regulator, showed an increased level of ompS2 expression. Augmented expression of ompS2 was also obtained upon addition of cloned leuO to the wild-type strain, but not in an ompR isogenic derivative, consistent with the notion that the transposon insertion had increased the cellular levels of LeuO and with the observed dependence on OmpR. Moreover, LeuO and OmpR bound in close proximity, but independently, to the 5' upstream regulatory region. Thus, the OmpR and LeuO regulators positively regulate ompS2.
Figures







References
-
- Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene 160:59-62. - PubMed
-
- Aron, L., G. Faundez, C. González, E. Roessler, and F. Cabello. 1993. Lipopolysaccharide-independent radio-immunoprecipitation and identification of structural and in vivo induced immunogenic surface proteins of Salmonella typhi in typhoid fever. Vaccine 11:10-17. - PubMed
-
- Benson, S. A., J. L. Occi, and B. A. Sampson. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K-12. J. Mol. Biol. 203:961-970. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

