Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi
- PMID: 15126483
- PMCID: PMC400632
- DOI: 10.1128/JB.186.10.3202-3213.2004
Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi
Abstract
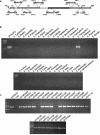
The large pathogenicity island (SPI7) of Salmonella enterica serovar Typhi is a 133,477-bp segment of DNA flanked by two 52-bp direct repeats overlapping the pheU (phenylalanyl-tRNA) gene, contains 151 potential open reading frames, and includes the viaB operon involved in the synthesis of Vi antigen. Some clinical isolates of S. enterica serovar Typhi are missing the entire SPI7, due to its precise excision; these strains have lost the ability to produce Vi antigen, are resistant to phage Vi-II, and invade a human epithelial cell line more rapidly. Excision of SPI7 occurs spontaneously in a clinical isolate of S. enterica serovar Typhi when it is grown in the laboratory, leaves an intact copy of the pheU gene at its novel join point, and results in the same three phenotypic consequences. SPI7 is an unstable genetic element, probably an intermediate in the pathway of lateral transfer of such pathogenicity islands among enteric gram-negative bacteria.
Figures



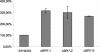

References
-
- Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

