-1 frameshifting at a CGA AAG hexanucleotide site is required for transposition of insertion sequence IS1222
- PMID: 15126494
- PMCID: PMC400620
- DOI: 10.1128/JB.186.10.3274-3277.2004
-1 frameshifting at a CGA AAG hexanucleotide site is required for transposition of insertion sequence IS1222
Abstract
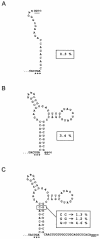
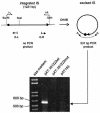
The discovery of programmed -1 frameshifting at the hexanucleotide shift site CGA_AAG, in addition to the classical X_XXY_YYZ heptanucleotide shift sequences, prompted a search for instances among eubacterial insertion sequence elements. IS1222 has a CGA_AAG shift site. A genetic analysis revealed that frameshifting at this site is required for transposition.
Figures

References
-
- Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2002. Recoding: translational bifurcations in gene expression. Gene 286:187-201. - PubMed
-
- Brierley, I., and S. Pennell. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic −1 frameshifting. Cold Spring Harbor Symp. Quant. Biol. 66:233-248. - PubMed
-
- Chandler, M., and O. Fayet. 1993. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7:497-503. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

