Simultaneous quantitative and allele-specific expression analysis with real competitive PCR
- PMID: 15128429
- PMCID: PMC411033
- DOI: 10.1186/1471-2156-5-8
Simultaneous quantitative and allele-specific expression analysis with real competitive PCR
Abstract
Background: For a diploid organism such as human, the two alleles of a particular gene can be expressed at different levels due to X chromosome inactivation, gene imprinting, different local promoter activity, or mRNA stability. Recently, imbalanced allelic expression was found to be common in human and can follow Mendelian inheritance. Here we present a method that employs real competitive PCR for allele-specific expression analysis.
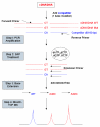
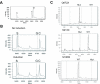
Results: A transcribed mutation such as a single nucleotide polymorphism (SNP) is used as the marker for allele-specific expression analysis. A synthetic mutation created in the competitor is close to a natural mutation site in the cDNA sequence. PCR is used to amplify the two cDNA sequences from the two alleles and the competitor. A base extension reaction with a mixture of ddNTPs/dNTP is used to generate three oligonucleotides for the two cDNAs and the competitor. The three products are identified and their ratios are calculated based on their peak areas in the MALDI-TOF mass spectrum. Several examples are given to illustrate how allele-specific gene expression can be applied in different biological studies.
Conclusions: This technique can quantify the absolute expression level of each individual allele of a gene with high precision and throughput.
Figures
References
-
- Bray NJ, Buckland PR, Owen MJ, O'Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

