GeneOrder3.0: software for comparing the order of genes in pairs of small bacterial genomes
- PMID: 15128433
- PMCID: PMC419981
- DOI: 10.1186/1471-2105-5-52
GeneOrder3.0: software for comparing the order of genes in pairs of small bacterial genomes
Abstract
Background: An increasing number of whole viral and bacterial genomes are being sequenced and deposited in public databases. In parallel to the mounting interest in whole genomes, the number of whole genome analyses software tools is also increasing. GeneOrder was originally developed to provide an analysis of genes between two genomes, allowing visualization of gene order and synteny comparisons of any small genomes. It was originally developed for comparing virus, mitochondrion and chloroplast genomes. This is now extended to small bacterial genomes of sizes less than 2 Mb.
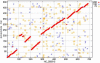
Results: GeneOrder3.0 has been developed and validated successfully on several small bacterial genomes (ca. 580 kb to 1.83 Mb) archived in the NCBI GenBank database. It is an updated web-based "on-the-fly" computational tool allowing gene order and synteny comparisons of any two small bacterial genomes. Analyses of several bacterial genomes show that a large amount of gene and genome re-arrangement occurs, as seen with earlier DNA software tools. This can be displayed at the protein level using GeneOrder3.0. Whole genome alignments of genes are presented in both a table and a dot plot. This allows the detection of evolutionary more distant relationships since protein sequences are more conserved than DNA sequences.
Conclusions: GeneOrder3.0 allows researchers to perform comparative analysis of gene order and synteny in genomes of sizes up to 2 Mb "on-the-fly."
Availability: http://binf.gmu.edu/genometools.html and http://pasteur.atcc.org:8050/GeneOrder3.0.
Figures
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous