Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster-type gene, ZFR1
- PMID: 15128515
- PMCID: PMC404460
- DOI: 10.1128/AEM.70.5.2653-2659.2004
Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster-type gene, ZFR1
Abstract
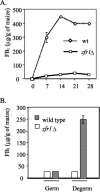
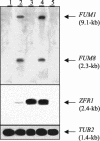
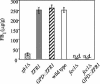
Fusarium verticillioides, a pathogen of maize, produces a class of mycotoxins called fumonisins in infected kernels. In this study, a candidate regulatory gene, ZFR1, was identified in an expressed sequence tag library enriched for transcripts expressed by F. verticillioides during fumonisin B(1) (FB(1)) biosynthesis. ZFR1 deletion mutants exhibited normal growth and development on maize kernels, but fumonisin production was reduced to less than 10% of that of the wild-type strain. ZFR1 encodes a putative protein of 705 amino acids with sequence similarity to the Zn(II)2Cys6 binuclear cluster family that are regulators of both primary and secondary metabolism in fungi. Expression of ZFR1 in colonized germ and degermed kernel tissues correlated with FB(1) levels. Overexpression of ZFR1 in zfr1 mutants restored FB(1) production to wild-type levels; however, FB(1) was not restored in an fcc1 (Fusarium C-type cyclin) mutant by overexpression of ZFR1. The results of this study indicate that ZFR1 is a positive regulator of FB(1) biosynthesis in F. verticillioides and suggest that FCC1 is required for ZFR1 function.
Figures



References
-
- Bibbins, M., V. F. Crepin, N. J. Cummings, T. Mizote, K. Baker, K. H. Mellits, and I. F. Connerton. 2002. A regulator gene for acetate utilisation from Neurospora crassa. Mol. Genet. Genomics 267:498-505. - PubMed
-
- Butchko, R. A., R. D. Plattner, and R. H. Proctor. 2003. FUM13 encodes a short chain dehydrogenase/reductase required for C-3 carbonyl reduction during fumonisin biosynthesis in Gibberella moniliformis. J. Agric. Food Chem. 51:3000-3006. - PubMed
-
- Cary, J. W., K. C. Ehrlich, M. Wright, P.-K. Chang, and D. Bhatnagar. 2000. Generation of AFLR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:680-684. - PubMed
-
- Chung, K. R., M. E. Daub, K. Kuchler, and C. Schuller. 2003. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 302:302-310. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

