Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense
- PMID: 15128517
- PMCID: PMC404397
- DOI: 10.1128/AEM.70.5.2667-2677.2004
Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense
Abstract
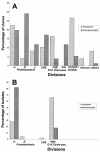
Thlaspi goesingense is able to hyperaccumulate extremely high concentrations of Ni when grown in ultramafic soils. Recently it has been shown that rhizosphere bacteria may increase the heavy metal concentrations in hyperaccumulator plants significantly, whereas the role of endophytes has not been investigated yet. In this study the rhizosphere and shoot-associated (endophytic) bacteria colonizing T. goesingense were characterized in detail by using both cultivation and cultivation-independent techniques. Bacteria were identified by 16S rRNA sequence analysis, and isolates were further characterized regarding characteristics that may be relevant for a beneficial plant-microbe interaction-Ni tolerance, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase and siderophore production. In the rhizosphere a high percentage of bacteria belonging to the Holophaga/Acidobacterium division and alpha-Proteobacteria were found. In addition, high-G+C gram-positive bacteria, Verrucomicrobia, and microbes of the Cytophaga/Flexibacter/Bacteroides division colonized the rhizosphere. The community structure of shoot-associated bacteria was highly different. The majority of clones affiliated with the Proteobacteria, but also bacteria belonging to the Cytophaga/Flexibacter/Bacteroides division, the Holophaga/Acidobacterium division, and the low-G+C gram-positive bacteria, were frequently found. A high number of highly related Sphingomonas 16S rRNA gene sequences were detected, which were also obtained by the cultivation of endophytes. Rhizosphere isolates belonged mainly to the genera Methylobacterium, Rhodococcus, and Okibacterium, whereas the majority of endophytes showed high levels of similarity to Methylobacterium mesophilicum. Additionally, Sphingomonas spp. were abundant. Isolates were resistant to Ni concentrations between 5 and 12 mM; however, endophytes generally tolerated higher Ni levels than rhizosphere bacteria. Almost all bacteria were able to produce siderophores. Various strains, particularly endophytes, were able to grow on ACC as the sole nitrogen source.
Figures

References
-
- Abeles, F. B., P. W. Morgan, and M. E. Saltveit, Jr. 1992. Regulation of ethylene production by internal, environmental and stress factors, p. 56-119. In Ethylene in plant biology, 2nd ed. Academic Press, Inc., San Diego, Calif.
-
- Abou-Shanab, R. A., J. S. Angle, T. A. Delorme, R. L. Chaney, P. van Berkum, H. Moawad, K. Ghanem, and H. A. Ghozlan. 2003. Rhizobacterial effects on nickel extraction from soil and uptake by Alyssum murale. New Phytol. 158:219-224.
-
- Baker, A. J. M., S. P. McGrath, R. D. Reeves, and J. A. C. Smith. 2000. Metal hyperaccumulator plants: a review of the ecology and physiology of a biological resource for phytoremediation of metal-polluted soils, p. 85-107. In N. Terry and E. Banuelos (ed.), Phytoremediation of contaminated soil and water. Lewis Publishers, London, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

