Effect of adaptation to ethanol on cytoplasmic and membrane protein profiles of Oenococcus oeni
- PMID: 15128528
- PMCID: PMC404408
- DOI: 10.1128/AEM.70.5.2748-2755.2004
Effect of adaptation to ethanol on cytoplasmic and membrane protein profiles of Oenococcus oeni
Abstract
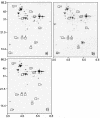
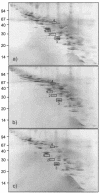
The practical application of commercial malolactic starter cultures of Oenococcus oeni surviving direct inoculation in wine requires insight into mechanisms of ethanol toxicity and of acquired ethanol tolerance in this organism. Therefore, the site-specific location of proteins involved in ethanol adaptation, including cytoplasmic, membrane-associated, and integral membrane proteins, was investigated. Ethanol triggers alterations in protein patterns of O. oeni cells stressed with 12% ethanol for 1 h and those of cells grown in the presence of 8% ethanol. Levels of inosine-5'-monophosphate dehydrogenase and phosphogluconate dehydrogenase, which generate reduced nicotinamide nucleotides, were decreased during growth in the presence of ethanol, while glutathione reductase, which consumes NADPH, was induced, suggesting that maintenance of the redox balance plays an important role in ethanol adaptation. Phosphoenolpyruvate:mannose phosphotransferase system (PTS) components of mannose PTS, including the phosphocarrier protein HPr and EII(Man), were lacking in ethanol-adapted cells, providing strong evidence that mannose PTS is absent in ethanol-adapted cells, and this represents a metabolic advantage to O. oeni cells during malolactic fermentation. In cells grown in the presence of ethanol, a large increase in the number of membrane-associated proteins was observed. Interestingly, two of these proteins, dTDT-glucose-4,6-dehydratase and D-alanine:D-alanine ligase, are known to be involved in cell wall biosynthesis. Using a proteomic approach, we provide evidence for an active ethanol adaptation response of O. oeni at the cytoplasmic and membrane protein levels.
Figures

References
-
- Arthur, M., P. E. Reynolds, F. Depardieu, S. Evers, S. Dutka-Malen, R. Quintiliani, and P. Courvalin. 1996. Mechanisms of glycopeptide resistance in enterococci. J. Infect. 32:10-16. - PubMed
-
- Bourassa, S., and C. Vadeboncoeur. 1992. Expression of an inducible enzyme II fructose and activation of a cryptic enzyme II glucose in glucose-grown cells of spontaneous mutants of Streptococcus salivarius lacking the low-molecular-mass of IIIman, a component of the phosphoenolpyruvate:mannose phosphotransferase system. J. Gen. Microbiol. 138:769-777. - PubMed
-
- Cavin, J. F., P. Schmitt, A. Arias, J. Lin, and C. Divies. 1988. Plasmid profiles in Leuconostoc species. Microbiol. Aliment. Nutr. 6:55-62.
-
- Chaillou, S., W. P. Postma, and P. H. Pouwels. 2001. Contribution of the phosphoenolpyruvate:mannose phosphotransferase system to carbon catabolite repression in Lactobacillus pentosus. Microbiology 127:671-679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

