Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae
- PMID: 15128548
- PMCID: PMC404438
- DOI: 10.1128/AEM.70.5.2892-2897.2004
Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae
Abstract
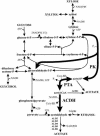
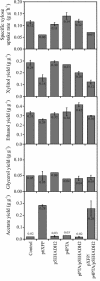
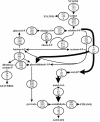
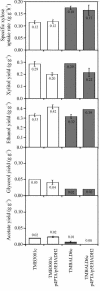
Low ethanol yields on xylose hamper economically viable ethanol production from hemicellulose-rich plant material with Saccharomyces cerevisiae. A major obstacle is the limited capacity of yeast for anaerobic reoxidation of NADH. Net reoxidation of NADH could potentially be achieved by channeling carbon fluxes through a recombinant phosphoketolase pathway. By heterologous expression of phosphotransacetylase and acetaldehyde dehydrogenase in combination with the native phosphoketolase, we installed a functional phosphoketolase pathway in the xylose-fermenting Saccharomyces cerevisiae strain TMB3001c. Consequently the ethanol yield was increased by 25% because less of the by-product xylitol was formed. The flux through the recombinant phosphoketolase pathway was about 30% of the optimum flux that would be required to completely eliminate xylitol and glycerol accumulation. Further overexpression of phosphoketolase, however, increased acetate accumulation and reduced the fermentation rate. By combining the phosphoketolase pathway with the ald6 mutation, which reduced acetate formation, a strain with an ethanol yield 20% higher and a xylose fermentation rate 40% higher than those of its parent was engineered.
Figures
Similar articles
-
Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose.Appl Environ Microbiol. 2002 Apr;68(4):1604-9. doi: 10.1128/AEM.68.4.1604-1609.2002. Appl Environ Microbiol. 2002. PMID: 11916674 Free PMC article.
-
Effects of NADH-preferring xylose reductase expression on ethanol production from xylose in xylose-metabolizing recombinant Saccharomyces cerevisiae.J Biotechnol. 2012 Apr 30;158(4):184-91. doi: 10.1016/j.jbiotec.2011.06.005. Epub 2011 Jun 15. J Biotechnol. 2012. PMID: 21699927
-
Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6.Microb Cell Fact. 2016 Apr 26;15:67. doi: 10.1186/s12934-016-0465-z. Microb Cell Fact. 2016. PMID: 27118055 Free PMC article.
-
[Progress in the pathway engineering of ethanol fermentation from xylose utilising recombinant Saccharomyces cerevisiae].Sheng Wu Gong Cheng Xue Bao. 2003 Sep;19(5):636-40. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15969099 Review. Chinese.
-
Engineering Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: reflections and perspectives.Biotechnol J. 2012 Jan;7(1):34-46. doi: 10.1002/biot.201100053. Epub 2011 Dec 7. Biotechnol J. 2012. PMID: 22147620 Review.
Cited by
-
Rewriting yeast central carbon metabolism for industrial isoprenoid production.Nature. 2016 Sep 29;537(7622):694-697. doi: 10.1038/nature19769. Epub 2016 Sep 21. Nature. 2016. PMID: 27654918
-
Metabolic engineering considerations for the heterologous expression of xylose-catabolic pathways in Saccharomyces cerevisiae.PLoS One. 2020 Jul 27;15(7):e0236294. doi: 10.1371/journal.pone.0236294. eCollection 2020. PLoS One. 2020. PMID: 32716960 Free PMC article.
-
A comparative analysis of NADPH supply strategies in Saccharomyces cerevisiae: Production of d-xylitol from d-xylose as a case study.Metab Eng Commun. 2024 Jul 5;19:e00245. doi: 10.1016/j.mec.2024.e00245. eCollection 2024 Dec. Metab Eng Commun. 2024. PMID: 39072283 Free PMC article.
-
Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae.mBio. 2014 Oct 21;5(5):e01696-14. doi: 10.1128/mBio.01696-14. mBio. 2014. PMID: 25336454 Free PMC article.
-
Genome-scale metabolic modeling reveals metabolic trade-offs associated with lipid production in Rhodotorula toruloides.PLoS Comput Biol. 2023 Apr 26;19(4):e1011009. doi: 10.1371/journal.pcbi.1011009. eCollection 2023 Apr. PLoS Comput Biol. 2023. PMID: 37099621 Free PMC article.
References
-
- Anderlund, M., P. Radstrom, and B. Hahn-Hägerdal. 2001. Expression of bifunctional enzymes with xylose reductase and xylitol dehydrogenase activity in Saccharomyces cerevisiae alters product formation during xylose fermentation. Metab. Eng. 3:226-235. - PubMed
-
- Aristidou, A., J. Londesborough, M. Penttilä, P. Richard, L. Rouhonen, H. Soderlund, A. Teleman, and M. Toivari. 1999. Transformed microorganisms with improved properties. Patent WO 99/46363.
-
- Bruinenberg, P. M., P. H. M. De Bot, J. P. van Dijken, and W. A. Scheffers. 1984. NADH-linked aldose reductase: the key to ethanolic fermentation of xylose by yeasts. Appl. Microbiol. Biotechnol. 19:256-260.
-
- Buu, L. M., Y. C. Chen, and F. J. Lee. 2003. Functional characterization and localization of acetyl-CoA hydrolase, Ach1p, in Saccharomyces cerevisiae. J. Biol. Chem. 278:17203-17209. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

